Details
-
Type:
New Feature
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: None
-
Labels:None
-
Story Points:1
-
Epic Link:
-
Sprint:Spring 8 : 24 Apr to 8 May, Spring 8 : 11 May to 25 May
Description
NCBI primer site has added a new parameter. To ensure IGB users have access to the newly available options, we should add this new option to NCBI Primer App.
See: https://ncbiinsights.ncbi.nlm.nih.gov/2020/04/24/primer-blast-3-match/
Attachments
Issue Links
- blocks
-
IGBF-2320 Add tooltips to NCBI Primer Design options panel
-
- Closed
-
Activity
| Field | Original Value | New Value |
|---|---|---|
| Epic Link |
|
| Rank | Ranked higher |
| Summary | Add new parameter | Add new parameter to NCBI Primer App |
| Assignee | Ann Loraine [ aloraine ] |
| Assignee | Noor Zahara [ noor91zahara ] |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Attachment | Screen Shot 2020-04-30 at 12.30.30 PM.png [ 14704 ] |
| Attachment | Screen Shot 2020-04-30 at 4.05.03 PM.png [ 14705 ] |
| Status | In Progress [ 3 ] | Needs 1st Level Review [ 10005 ] |
| Assignee | Noor Zahara [ noor91zahara ] |
| Sprint | Spring 8 : 24 Apr to 8 May [ 93 ] | Spring 8 : 24 Apr to 8 May, Spring 8 : 11 May to 25 May [ 93, 94 ] |
| Rank | Ranked higher |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Assignee | Pooja Nikhare [ pooja.nikhare ] |
| Status | First Level Review in Progress [ 10301 ] | Ready for Pull Request [ 10304 ] |
| Assignee | Pooja Nikhare [ pooja.nikhare ] | Noor Zahara [ noor91zahara ] |
| Status | Ready for Pull Request [ 10304 ] | Pull Request Submitted [ 10101 ] |
| Assignee | Noor Zahara [ noor91zahara ] |
| Status | Pull Request Submitted [ 10101 ] | Reviewing Pull Request [ 10303 ] |
| Status | Reviewing Pull Request [ 10303 ] | Merged Needs Testing [ 10002 ] |
| Status | Merged Needs Testing [ 10002 ] | Post-merge Testing In Progress [ 10003 ] |
| Resolution | Done [ 10000 ] | |
| Status | Post-merge Testing In Progress [ 10003 ] | Closed [ 6 ] |
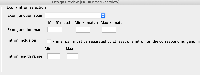
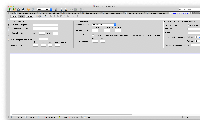
I have designed the GUI for the new parameter. I have attached. Let me know if it looks fine.
I had a doubt. I do need to add the entire block that I have attached right?