I have tested each quickload file in the 2 scenarios.
When I tested in the first scenario using H_sapiens_Dec_2013 human genome, part of the data was not loaded and mostly they are Soft-clips.
I have added 5 images for that scenario and log for the error is:
15:31:13.752 WARN o.l.i.quickload.util.QuickloadUtils - Optional species.txt could not be loaded from: https://bitbucket.org/nfreese/quickload-genome-in-a-bottle/raw/release-v1.0.1/quickload/species.txt
15:31:13.826 INFO o.l.i.appstore.IgbAppServerLauncher - Started REST endpoint.
When I tested in the first scenario using H_sapiens_Dec_2009 human genome, this issue is not there but in AshkenazimTrio --> Son --> Illumina_6kb_matepair was not loaded and log for the error is:
17:31:38.694 INFO c.a.igb.view.load.GeneralLoadUtils - Loaded RefGene in 2.188 s
org.lorainelab.igb.bam.BAM$BamIndexNotFoundException: Could not find Bam Index File.
at org.lorainelab.igb.bam.BAM.findIndexFile(BAM.java:433)
at org.lorainelab.igb.bam.BAM.getBamIndexUriStr(BAM.java:139)
at org.lorainelab.igb.bam.BAM.getSAMFileReader(BAM.java:98)
at org.lorainelab.igb.bam.BAM.init(BAM.java:157)
at org.lorainelab.igb.bam.XAM.getChromosomeList(XAM.java:100)
at com.affymetrix.genometry.quickload.QuickLoadSymLoader.loadAndAddSymmetries(QuickLoadSymLoader.java:153)
at com.affymetrix.genometry.quickload.QuickLoadSymLoader.loadSymmetriesThread(QuickLoadSymLoader.java:139)
at com.affymetrix.genometry.quickload.QuickLoadSymLoader.loadFeatures(QuickLoadSymLoader.java:119)
at com.affymetrix.igb.view.load.GeneralLoadUtils.loadFeaturesForSym(GeneralLoadUtils.java:751)
at com.affymetrix.igb.view.load.GeneralLoadUtils$2.runInBackground(GeneralLoadUtils.java:693)
at com.affymetrix.igb.view.load.GeneralLoadUtils$2.runInBackground(GeneralLoadUtils.java:688)
at com.affymetrix.genometry.thread.CThreadWorker.doInBackground(CThreadWorker.java:73)
at javax.swing.SwingWorker$1.call(SwingWorker.java:295)
at java.util.concurrent.FutureTask.run(FutureTask.java:266)
at javax.swing.SwingWorker.run(SwingWorker.java:334)
at java.util.concurrent.ThreadPoolExecutor.runWorker(ThreadPoolExecutor.java:1149)
at java.util.concurrent.ThreadPoolExecutor$Worker.run(ThreadPoolExecutor.java:624)
at java.lang.Thread.run(Thread.java:748)
cc: Dr.Nowlan Freese
Improvement
Major
IGBF-1217 add Genome in a Bottle data sets to IGB Quickload

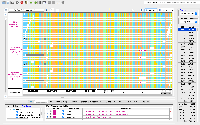


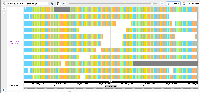

To test in IGB:
Add this link as a new Quickload under the Data Access tab in Available Data - Configure:
https://bitbucket.org/nfreese/quickload-genome-in-a-bottle/raw/release-v1.0.1/quickload/
Open the H_sapiens_Dec_2013 human genome.
Navigate to chr1:80,644,218-80,644,359
Select one of the Quickload files and click Load Data.
Look to see that the data load and no errors are thrown.
Repeat for every Quickload file.
Open the H_sapiens_Feb_2009 human genome.
Navigate to chr1:77,876,660-77,876,761
Select one of the Quickload files and click Load Data.
Look to see that the data load and no errors are thrown.
Repeat for every Quickload file.