Details
-
Type:
Improvement
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: 9.1.8 Major Release
-
Labels:None
-
Story Points:2
-
Epic Link:
-
Sprint:Winter 4 Feb 8 - Feb 19, Winter 5 Feb 22 - Mar 5, Winter 6 Mar 8 - Mar 19, Spring 1 2021 Mar 22 - Apr 2, Spring 2 2021 Apr 5 - Apr 16, Spring 3 2021 Apr 19 - Apr 30
Description
Situation: The Open Genome from File user interface can be improved.
Task: Improve the Open Genome from File user interface.
For example:
1) Genome Version should be below Species to make it more in line with the Species menu.
2) The placeholder for Species should be changed to something to more accurately reflect the expected input, such as "Genus species"
3) Change the Genome Version from a text box to a series of dropdowns. The Genome Version in IGB must conform to the following pattern:
H_sapiens_Dec_2013
We can obtain the first half by using the Genus species from the Species input. So if the user enters Homo sapiens, we would take the first letter of Homo (H) and then the second word (sapiens), separating them by underscores. We could then have a dropdown for the month (Jan/Feb/Mar/etc...) and then an input for year. We would then use these values to construct the IGB compliant Genome Version.
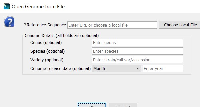
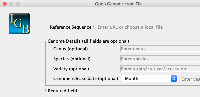
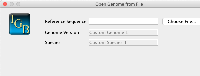
On selection of the species, the first word of the species name is abbreviated and the second word is added separated by an underscore for the version name. The current month and year is added to the name as well (separated by underscores). To change the version, the user can select the month from dropdown to change the month in the version name. The user can manually edit the year and the rest of the name within the genome version text field.