Details
-
Type:
Improvement
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: 9.1.8 Major Release
-
Labels:None
-
Story Points:2
-
Epic Link:
-
Sprint:Winter 4 Feb 8 - Feb 19, Winter 5 Feb 22 - Mar 5, Winter 6 Mar 8 - Mar 19, Spring 1 2021 Mar 22 - Apr 2, Spring 2 2021 Apr 5 - Apr 16, Spring 3 2021 Apr 19 - Apr 30
Description
Situation: The Open Genome from File user interface can be improved.
Task: Improve the Open Genome from File user interface.
For example:
1) Genome Version should be below Species to make it more in line with the Species menu.
2) The placeholder for Species should be changed to something to more accurately reflect the expected input, such as "Genus species"
3) Change the Genome Version from a text box to a series of dropdowns. The Genome Version in IGB must conform to the following pattern:
H_sapiens_Dec_2013
We can obtain the first half by using the Genus species from the Species input. So if the user enters Homo sapiens, we would take the first letter of Homo (H) and then the second word (sapiens), separating them by underscores. We could then have a dropdown for the month (Jan/Feb/Mar/etc...) and then an input for year. We would then use these values to construct the IGB compliant Genome Version.
Attachments
- Completed.jpg
- 36 kB
Issue Links
Activity
Genus: textbox (default value is Genus) only allow a-z and A-Z and no whitespace
Species: textbox (default value is species) only allow a-z and A-Z and no whitespace
Month: dropdown (default value is MMM)
Year: four numbers (default is YYYY)
Genome Version: updates dynamically but is not editable by the user
The getSpeciesName() public method is called from various places in the IGB code and so we need to make sure it returns the expected value. The value that should be returned is the combined "Genus species". So if the user entered "Homo" for genus and "sapiens" for species, the getSpeciesName() method should return "Homo sapiens".
Fixed the getSpeciesName() method return value.
1) There was an error where the users cannot change focus from a text field if there is not value or it the value is erroneous. That is fixed.
2) As discussed, placeholders has been added for all text fields if the user chooses to enter one or more values. Currently the logic is that the user can choose not to fill any fields and hit 'Ok' and the 'Genome Version' will be 'Custom Genome 1'. If any of the fields has value, it replaces the placeholders in 'Genus_species_MMM_YYYY' format, for example, if the user adds 2021 in the year field, the version name will be 'Genus_species_MMM_2021'.
3) A window height problem was also observed, 'Genome Version' name field is cut-off by the fixed height. The problem was handled by repositioning the month and year fields to fill the same cell in the layout thus eliminating the need for one more row in the layout.
Request for Karthik Raveendran: Please edit the previous comment? I cannot understand your meaning.
Based on this page of the user's guide we should probably include an optional Variety text box. Include a comment to the side of the text box that states "Optional - strain/cultivar/accession". Do not format the text in the Variety text box. The Variety text box can include uppercase/lowercase letters/numbers. Do not include any default text if the user leaves the Variety text box empty. A completed Genome Version with the optional Variety text would appear as: E_unicornis_un1c02n_Jul_2043
For the Genus text box, can we format the text so that the first letter is uppercase and the rest of the letters are lowercase?
Two changes:
Need to include varietyTextField as part of species name (line 267 I think).
Need additional vertical space in Open Genome from File box to keep bottom from being cut off. Or we could just not show the auto-completed Genome Version field, since the users can't interact with it.
Changes are implemented and pushed. The two images attached to this issue, Completed and Completed Validation, shows the new form window and the form after validation errors are caught.
It is not clear from the interface what the user is supposed to do. Some issues:
- As per discussion, "Version" appears to be a text entry box, but it is not editable.
- There is no way for a user to know what date they are supposed to enter. Should it be today's date? Some other date?
- It is not clear which fields are absolutely required and which are optional.
- This form assumes that a user is already familiar with IGB genome version syntax. A power user might be, but probably not.
Suggestion:
Create sample forms using a design tool and test it with user representatives to determine if it makes sense. Review chapters on usability testing in "A Practical Guide to Usability Testing". See: https://www.dropbox.com/sh/vqqznxr8xox56qg/AABIbGM46mAallhox70WiduHa
Usability test observations are in the link below:
https://docs.google.com/document/d/1ZaTt30LMdfk97480zlReLXG1bG8rjr3NPP3mOw8xtuU/edit?usp=sharing
The layout looks really good on Mac.
I wanted to make some changes to the getSpeciesName logic so I made a branch with some edits. I also removed the red boxes. I like the idea of them, but I think they are too much work for how useful they would be. We would need to add tooltips to inform the user why the text they have entered is problematic. And then I started thinking that there may be some species names with non-english characters, such as "é".
Check out my branch and copy over the changes if they look good to you (I'm not sure if it would be easier to just copy/paste or merge onto your branch).
https://bitbucket.org/nfreese/nowlanfork-igb/branch/IGBF-2783-NOWLAN
Next step: Karthik is working on fixing his bitbucket pipelines. Once fixed, it would be good to have someone pull down the installer and test before submitting a pull request.
The build pipeline is fixed. The installer was tested by me and Omkar Marne. Pull request has been submitted.
The design is better, but there are still some aspects that are confusing to me:
- Why is there an asterisk in front of "Reference Sequence" - I'm not sure what this is trying to communicate.
- When I clicked "Open File", a file chooser opened with a menu containing empty parentheses and the word Sequence file
- Also, when I did the above, I got a little lost because there were two modal dialogs open at the same time. I lost track of the second one on my desktop and it was confusing to me because I could no longer do anything with the other two windows that were already open.
- How does a user know when they should use the "Open Genome from File" option? The interface currently does not explain this.
- Why should I fill out the Genome Details if all fields are optional? What benefit do I get from doing this? Can I change these values later?
Also it is possible to open multiple file choosers.
Asterisk was added due to user feedback testing - the user's felt that an asterisk indicated that a field was required. We can remove it if confusing.
We would need to update the user's guide to explain the new UI and detail what the Genome Details fields will change in IGB.
I'm not sure that the remaining issues are within the scope of this ticket.
Change "Choose Local File..." to "Browse"
Move asterisk to right side "Reference Sequence*" and make the * red.
Add "*Required field" at the bottom of the modal, and make the * red.
Add description to bottom: "If you include the optional details IGB will display the details in the species and genome menus."
Karthik Raveendran - can you increase the width and height of the custom genome modal, the side and bottom are being cut off just a little bit on Mac (image attached).
Otherwise looks good.
Width/height look good on Mac.
Recommend pull request. (please squash commits)
Post-merge testing on Windows 10.
Everything looks right and functions correctly when I put the info in correctly.
When I enter an input for the Year that is not exactly four numbers, like '20211' or '202', the genome version defaults to "Custom Genome 1", ignoring all of the other fields I filled in to the form. Also, it lets me type in any amount of letter or number characters into the Year field without letting me know that it will cause a problem.
The other text entry fields allow me to use all number characters if I want.
If I fill in all of the fields except for the Species field, then it will generate a "Custom Species" text for that field in IGB.
In all other arrangements of leaving fields blank nothing appears in the Species field in IGB.
Would we want that default value to appear in every case ?
What do you think, Karthik Raveendran?
Is this happening on other machines?
In the first case, regarding the year field, when the user adds any erroneous input in any of the fields we revert to the default "Custom Genome" name for the genome version so that we don't restrict the user's movement in the application with error messages because these fields are optional. While we can add an upper limit (4) to the number of characters in that field, adding a lower limit (4) would require displaying an error message when the user enters input such as "202".
Regarding the other fields, the species field in IGB will only have values if the Genus and Species fields in the dialog window are filled out. The logic for that is left unchanged from the previous versions.
I spoke with Karthik and it looks like all of the behaviors I observed per my previous comment are normal and intended.
I have one question I wanted to run by Nowlan Freese,
So we want to make sure the user doesn't have to deal with a bunch of error messages for optional fields, but on the flip side -
Isn't it confusing if we forgo giving the user any error feedback when they do something that will result in the software not behaving as expected, like dropping all of their input data without warning?
Change request Karthik Raveendran:
1) If all fields are left blank the text should be generated: "Custom Species 1" and "Custom Genome 1" (incrementing the numbers where appropriate) for the Species and Genome Version menus within the Current Genome tab in IGB.
2) Limit the Year field to number characters, and/or take only the first 4 characters entered.
Question for Karthik Raveendran & Logan Weidenhammer:
- What do you think of this idea?
When the user first sees the form, the year and month are already filled in using the current year and month.
[~aloraine], If the Month and Year fields are already entered and the user does not enter any other values, then the "Custom Genome 1" format would never appear. It will always be in the format "Genus_Species_Apr_2021". With the current logic, if the user enters two reference seqs with the same name (and without any other optional entries), there would not be a clash in names because the "Current Genome" format increments the numbers. If there is a clash, both sequences will be merged together as one in IGB.
Logan Weidenhammer & Nowlan Freese, ill make both the changes.
Success:
All fields left blank works.
Year field works.
Issue:
Leaving only the Species field blank causes the Species dropdown in IGB to populate with the text from the Variety field.
If no Variety text entered from user, Species in IGB is shown as "Custom Species 1".
Genus should not be forced lowercase in text field
Variety should be appended to the species Name dropdown value
If the species field is left empty but variety is filled out, the species dropdown does not include the genus
If user enters only Genus then make the Species dropdown, for example:
Homo SPECIES
GENUS sapiens
GENUS SPECIES userEnteredVarietyValue
Default values should be all uppercase (GENUS SPECIES MMM YYYY) - continue to omit default values for variety since they are rarely used
Change requests mentioned above have been implemented
Rachel and I did additional testing after Karthik made some changes.
Recommend pull request.
Please bring the branch up-to-date with the latest master branch and check functionality.
PR was submitted and merged. I see this has not yet exited first level review so I'm not sure if I should have done that. My apologies.
Nowlan Freese and Karthik Raveendran - can you check it?
Rachel and I tested on Windows and Mac. Working well.
Closing issue.
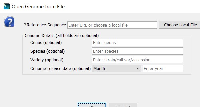
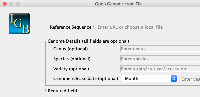
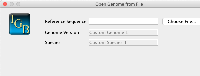
On selection of the species, the first word of the species name is abbreviated and the second word is added separated by an underscore for the version name. The current month and year is added to the name as well (separated by underscores). To change the version, the user can select the month from dropdown to change the month in the version name. The user can manually edit the year and the rest of the name within the genome version text field.