Details
-
Type:
Improvement
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: None
-
Labels:None
-
Story Points:4
-
Epic Link:
-
Sprint:Winter 6 Mar 8 - Mar 19, Spring 1 2021 Mar 22 - Apr 2, Spring 2 2021 Apr 5 - Apr 16, Spring 3 2021 Apr 19 - Apr 30
Description
Situation: The Quickload Saver app outputs an IGB Quickload. It does not currently include the optional species.txt file.
Task: Output the species.txt file when using the Quickload Saver app.
Attachments
- NewOpenGenome.PNG
- 18 kB
Issue Links
- relates to
-
IGBF-510 Create Quickload Saver IGB app
-
- Closed
-
Activity
Look at how Noor outputs the contents.txt. You can use similar code to output the species.txt.
Working on adding species.txt section to the QuickloadSaver App code.
On bitbucket, I see my topic branch on my remote fork and it does build in pipelines.
But it doesn't produce any files in my downloads section.
I can't see this branch in my branches section on Bitbucket.
I created a new branch from the master locally, then pushed the changes to my remote repo.
I need to see if this is what's causing the files to not appear in my downloads.
Thank you to Chester.
The App Password and Repository Variables were not set up before.
The Pipelines are now properly building the jar and xml files in the Downloads section.
Fork link for reference: https://bitbucket.org/Rweidenh/quickload-saver/downloads/
I haven't pushed anything remotely at this time.
I am still working on implementing the species.txt file creation ability in the app's code.
What's slowing me down right now is trying to understand the object classes and their function.
I am looking for a javadoc type documentation.
For example:
genomeVersion, getSpeciesName() getDescription()
What information is stored here exactly, where is it taken from, and what class do these come from?
Working on getting a plain species.txt file to be created.
I am starting to understand the existing code.
Altered some hard coding to accommodate an additional file.
I am trying to understand now why the annots file code looks to have a different way of creating a file than the contents file does.
Edit: The code for creating files appeared to be labeled as code for just the contents.txt file, but it was actually a new section of its own, responsible for creating all files previous to it.
All files are now being created, and in the right place too.
The information needed for species.txt is in the file.
Now I need to clean up the formatting and separate out the exact information we need.
I have pushed my local changes to my remote branch for reference.
The file is being created and the main content: the latin name, common name, and version prefix, are being created in the file.
The "Galaxy/UCSC genome version prefix" is not created but I'm not sure if we wanted to have that function here.
Looking at it now, my current logic for getting the information for the common name and the version prefix from the genome version description and name will not be able to handle common names with more than one word or version prefixes with more than two parts.
I will have to rethink my solution to this.
Maybe I can get to where the genome version description and genome version name are being created, but that location is not the GenomeVersion.java file.
Link to fork downloads page: https://bitbucket.org/Rweidenh/quickload-saver/downloads/
Link to branch: https://bitbucket.org/Rweidenh/quickload-saver/branch/IGBF-2804
There are four columns to a Species.txt file:
Latin Name, Common Name, Genome Prefix, and Genome Prefixes used by other data providers (UCSC/Galaxy).
For the purposes of the Quickload Saver app, Dr. Freese and I have decided to have the app only populate the first and third (Latin Name and Genome Prefix) columns, with the Common Name column being replaced by a TAB character. The idea is that this covers the basics of what IGB needs to populate the "Species" and "Genome Version" dropdown menus.
More detail - test using a species and genome that do not already exist in the species and genome version menus.
To Test:
Follow the instructions here: https://wiki.transvar.org/display/ITD/Quickload+Saver
Don't use the LoraineLab link in the instructions (https://bitbucket.org/lorainelab/quickload-saver/downloads/)
Instead use the topic branch link: https://bitbucket.org/Rweidenh/quickload-saver/downloads/
After completing the wiki instructions, find the quickload files you created and saved to your local machine.
Open the files and find the species.txt file created.
In the species.txt file you should see the Latin Name (e.g. equus unicornis), followed by three TAB spaces, followed by the Genome Prefix (e.g. E_unicornis).
Can confirm, followed these steps on the latest 9.1.8 installer and in the species.txt file, I am indeed seeing the latin name, the three tab spaces and the genome prefix. Everything looks good! Recommending pull request
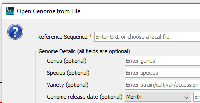
Moved back to needs post-merge testing. Quickload saver does indeed output correct species.txt file contents when a quickload is saved. However, it only does so whenever optional genome details (i.e. Genus, Species, Variety, Genome release date) from Karthik's recent UI change (see screenshot) are filled in.
Otherwise, if those fields are blank, there have been two different types of observed results taking place: either the quickload won't save anything to the user-specified folder or the quickload will save but will only include the annots.xml file.
Plan is to test again once all various code changes are merged to master, just before release testing.
Pasted from log after no files were saved to new quickload folder from blank optional fields:
10:57:56.542 INFO c.a.igb.view.load.GeneralLoadUtils - Loaded E_unicornis_Jul_2043.bed.gz in 37.22 ms
Exception in thread "AWT-EventQueue-0" java.lang.ArrayIndexOutOfBoundsException: 1
at org.lorainelab.igb.menuoption.CreateQuickloadAction.createQuickload(CreateQuickloadAction.java:133)
at org.lorainelab.igb.menuoption.CreateQuickloadAction.showFileChooser(CreateQuickloadAction.java:104)
at org.lorainelab.igb.menuoption.CreateQuickloadAction.actionPerformed(CreateQuickloadAction.java:75)
at org.lorainelab.igb.menuoption.CreateQuickloadAction.lambda$getMenuItems$0(CreateQuickloadAction.java:301)
at org.lorainelab.igb.menu.api.util.MenuUtils$1.actionPerformed(MenuUtils.java:26)
at javax.swing.AbstractButton.fireActionPerformed(Unknown Source)
at javax.swing.AbstractButton$Handler.actionPerformed(Unknown Source)
at javax.swing.DefaultButtonModel.fireActionPerformed(Unknown Source)
at javax.swing.DefaultButtonModel.setPressed(Unknown Source)
at javax.swing.AbstractButton.doClick(Unknown Source)
at javax.swing.plaf.basic.BasicMenuItemUI.doClick(Unknown Source)
at javax.swing.plaf.basic.BasicMenuItemUI$Handler.mouseReleased(Unknown Source)
at java.awt.Component.processMouseEvent(Unknown Source)
at javax.swing.JComponent.processMouseEvent(Unknown Source)
at java.awt.Component.processEvent(Unknown Source)
at java.awt.Container.processEvent(Unknown Source)
at java.awt.Component.dispatchEventImpl(Unknown Source)
at java.awt.Container.dispatchEventImpl(Unknown Source)
at java.awt.Component.dispatchEvent(Unknown Source)
at java.awt.LightweightDispatcher.retargetMouseEvent(Unknown Source)
at java.awt.LightweightDispatcher.processMouseEvent(Unknown Source)
at java.awt.LightweightDispatcher.dispatchEvent(Unknown Source)
at java.awt.Container.dispatchEventImpl(Unknown Source)
at java.awt.Window.dispatchEventImpl(Unknown Source)
at java.awt.Component.dispatchEvent(Unknown Source)
at java.awt.EventQueue.dispatchEventImpl(Unknown Source)
at java.awt.EventQueue.access$500(Unknown Source)
at java.awt.EventQueue$3.run(Unknown Source)
at java.awt.EventQueue$3.run(Unknown Source)
at java.security.AccessController.doPrivileged(Native Method)
at java.security.ProtectionDomain$JavaSecurityAccessImpl.doIntersectionPrivilege(Unknown Source)
at java.security.ProtectionDomain$JavaSecurityAccessImpl.doIntersectionPrivilege(Unknown Source)
at java.awt.EventQueue$4.run(Unknown Source)
at java.awt.EventQueue$4.run(Unknown Source)
at java.security.AccessController.doPrivileged(Native Method)
at java.security.ProtectionDomain$JavaSecurityAccessImpl.doIntersectionPrivilege(Unknown Source)
at java.awt.EventQueue.dispatchEvent(Unknown Source)
at java.awt.EventDispatchThread.pumpOneEventForFilters(Unknown Source)
at java.awt.EventDispatchThread.pumpEventsForFilter(Unknown Source)
at java.awt.EventDispatchThread.pumpEventsForHierarchy(Unknown Source)
at java.awt.EventDispatchThread.pumpEvents(Unknown Source)
at java.awt.EventDispatchThread.pumpEvents(Unknown Source)
at java.awt.EventDispatchThread.run(Unknown Source)
Three issues:
Quickload Saver index out of bounds
Quickload Saver not saving foreground color
Quickload Saver extra tab in species.txt
Extra tab is removed from species.txt
Index out of bounds exception no longer present and files are created appropriately.
If the user opens a new genome from file and enters no data, the species.txt file will show:
Custom Species 1 (TAB) (TAB) Custom Genome 1
Working on foreground color saving now.
Logan Weidenhammer - I fixed the foreground color, it was related to the change I made to the URI encoding.
Rachel and I tested on Windows and Mac. Working correctly.
Closing issue.
Quickload Saver: https://bitbucket.org/lorainelab/quickload-saver/src/master/
Species.txt documentation: https://wiki.transvar.org/display/igbman/Sharing+data+for+a+custom+genome+not+already+part+of+IGB+QuickLoad#SharingdataforacustomgenomenotalreadypartofIGBQuickLoad-Createspecies.txtfile