Details
-
Type:
Task
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: 10.0.0 Major Release
-
Labels:None
-
Story Points:5
-
Sprint:Spring 7, Spring 9, Summer 1, Summer 2, Summer 3, Summer 4, Summer 5
Description
Bob Goldstein, faculty from UNC Chapel Hill, is coming to Charlotte to give a seminar on Friday April 5 at 2:30.
His research Web site: https://goldsteinlab.weebly.com/
His tardigrades site: http://tardigrades.bio.unc.edu/
Ann Loraine signed up for a meeting with him at 1:30 in the third floor conference room. (We might use my office instead, depending on whether they finished repairing the walls yet - long story!)
Our goal is to talk with him about the tardigrade genome project and how we can represent tardigrade (water bears) in IGB.
Water bears are an emerging model system for studying how animals survive in extreme environments.
According to the UCSC Genome Browser, they now have three tardigrade genome assemblies available.
Bob Goldstein's tardigrades web site (http://tardigrades.bio.unc.edu/) mentions that they are "developing the water bear Hypsibius exemplaris as a new model for studying evo-devo and resistance to extremes."
Their tardigrade site has a "links" page that includes a blast search link hosted at NCBI. That page allows users to search genome assembly
"Hypsibius dujardini GenBank assembly GCA_002082055.1"
UCSC appears to also feature this same genome assembly on their site. They call it "nHd_3.1 Apr. 2017 H.dujardini (Z151 2017 tardigrades) (GCA_002082055.1)"
See:
For this task:
- Get reference 2bit genome sequence file for the above genome assembly
- Investigate and get reference gene model annotations for the above assembly
- Add reference genome assembly to IGB Quickload main
- Add reference gene model annotations to IGB Quickload main
- Review Goldstein lab publications to identify foundational RNA-Seq data for this organism
Let's aim to get the tardigrade data added to IGB QL before Friday so that we can:
- show Dr. Goldstein an IGB demo with the tardigrade genome data
- ask Dr. Goldstein which RNA-Seq data sets are most relevant to his research
- tentatively schedule an on-line demo of IGB with Dr. Goldstein lab members
Attachments
Issue Links
Activity
Hypsibius tardigrade:
https://www.ncbi.nlm.nih.gov/datasets/genome/?taxon=58670
GCA_002082055.1 - Hypsibius exemplaris > https://www.ncbi.nlm.nih.gov/datasets/genome/GCA_002082055.1/
GCA_001579985.1 - Hypsibius dujardini > https://www.ncbi.nlm.nih.gov/datasets/genome/GCA_001579985.1/
GCA_001455005.1 - Hypsibius dujardini > https://www.ncbi.nlm.nih.gov/datasets/genome/GCA_001455005.1/
Note that the submitter for GCA_001455005.1 is Bob Goldstein, but NCBI warns that this genome assembly has been contaminated.
The genome assembly contains sequences from other organisms, cloning vectors, linkers, adapters, or primers.
It looks like only Hypsibius exemplaris and Ramazzottius varieornatus have genome annotations available.
Based on this paper it looks like they are interested in using Hypsibius exemplaris as the model.
Ann Loraine - Here are the files for Hypsibius exemplaris: https://drive.google.com/drive/folders/1Yc5eNuJcMTW1b1Q7wOrPa4JQXR-XR4W0?usp=sharing
- GCA_002082055.1_nHd_3.1_genomic.2bit
- GCA_002082055.1_nHd_3.1_genomic.gff.gz
- genome.txt
Thanks Nowlan Freese! Please delete that google folder because I have what I need!
I have added the genome to the svn repository and updated the quickload "main" content deployed at these locations:
To test:
- Start IGB
- Use Species and Genome menus (Current Genome tab) to select the new genome H_exemplaris_Z151_Apr_2017 for species Hypsibius exemplaris strain Z151
- Check that the full species name appears in the species menu with tooltip "water bear"
- All the gene models should load automatically. I have configured them so that the description field appears as the names.
- Check that the Web site content makes sense. Visit the first URL above and scroll down the page to check that there is an entry for the new genome in a similar style to all the others
- Enter the genome directory folder for "water bear" and check that the information there in the "header.md" file appearing above the files listing makes sense. If it doesn't, make a note here and suggest a fix.
Note that I converted the GFF file from GenBank to bed-detail using a newly committed version of this script: gff3ToBedDetail.py on branch "master" in repository https://bitbucket.org/lorainelab/genomesource
How to update the genome dashboard with common name and image for the water bear?
Looks like we would add the image in this folder with the name H_exemplaris_Z151.jpg
Based on this code from the genome dashboard, it looks like it is pulling the species.txt and using that to determine the common names.
To make sure it gets added to genome dashboard in a nice way:
- Clone https://bitbucket.org/lorainelab/genome-dashboard/
- Add image to public/images
- Edit species.txt and synonyms.txt for IGB release branch (genome dashboard retrieves these files from the branch)
- Make sure that the new genome is available from the primary Quickload main site used by that branch (see preferences json file for the release branch)
Note that genome dashboard, when deployed, will be configured to use the currently released IGB branch code base to coordinate between IGB and the quickload sites it knows about.
To do this for tardigrade, I made a branch with the required changes and did a PR from my fork and branch to the main branch on the team repository.
PR to update genome dashboard util.js to point at the main IGB branch: https://bitbucket.org/lorainelab/genome-dashboard/pull-requests/29
Commit: https://bitbucket.org/nfreese/genome-dashboard/commits/d66c03a40668bb19466143edf3cc5462011e341f
Nowlan Freese has submitted PR to update the default IGB branch for util.js in genome-dashboard code to "main" instead of "master"
Reviewing:
- Genome Dashboard is working correctly, shows common and scientific name, clicking on it opens the correct genome in IGB.
- Hypsibius exemplaris showing correctly in IGB. Annotations are loading by default, annotations look the same when comparing them to UCSC.
Only question I have, the annots.xml lists the title as "Annotation submitted by Keio University (March 3, 2022)", which then shows as the track name. Was the intention to show it as the track name? I was also a little confused as the listed submission date is March 4, 2022.
To do:
Fix typo in annots.xml - change March 3, 2022 -> March 4, 2022PK committed this change via svn on 4/24/24Add R. varieornatus tardigrade genome and annotation to IGB Quickload.- Update gff3ToBedDetail.py in GenomeSource repo so that it handles non protein coding genes.
R. varieornatus : https://www.ncbi.nlm.nih.gov/datasets/genome/GCA_001949185.1/
All the taxon tardigrada genome assemblies:
Ann Loraine - I have added the files for Ramazzottius varieornatus to Google Drive: https://drive.google.com/drive/folders/1acW5w8dMGvxit3kw9i_jiW9ZOUqV6W8X?usp=drive_link
- GCA_001949185.1_Rvar_4.0_genomic.2bit
- GCA_001949185.1_Rvar.gff
Files are from NCBI: https://www.ncbi.nlm.nih.gov/datasets/genome/GCA_001949185.1/
When I tried to run the latest version of gff3ToBedDetail.py I was seeing several warnings, so I was hoping you could take a look.
Update:
- Publication date for R. varieornatus assembly: 20 September 2016
- Strain is YOKOZUNA-1 according to Genbank record at https://www.ncbi.nlm.nih.gov/datasets/genome/GCA_001949185.1
- Therefore genome version should be added to IGB as: R_varieornatus_YOKOZUNA-1_Sep_2016
- Genome version synonyms: Rvar_4.0, GCA_001949185.1
Investigating GFF file from NCBI to try to understand errors noted by Nowlan Freese:
example line:
BDGG01000001.1 DDBJ CDS 441 704 . + 2 ID=cds-GAU87103.1;Parent=gene-RvY_00001;Dbxref=NCBI_GP:GAU87103.1;Name=GAU87103.1;gbkey=CDS;gene=RvY_00001-1;locus_tag=RvY_00001;product=hypothetical protein;protein_id=GAU87103.1
This feature type is "CDS" indicating that this line specifies a region of genomic sequence that is translated into protein. The "parent" identifier is "gene-RvY_00001". This means that anything with this same "parent" is part of the same gene model, usually. However, it's a bit weird because usually "gene" means: a collection of gene models that are theoretically produced by alternative transcription.
This means that the parser we write for this file needs to use the "parent" identifier to assemble components of a gene model.
The GFF "extra feature" section (the final column) also reports that the CDS has a "locus_tag" which seems like it could indicate a grouping that I normally think of as a "gene."
Searching for the parent identifier using grep brings up:
local aloraine$ grep RvY_00001-1 GCA_001949185.1_Rvar.gff BDGG01000001.1 DDBJ gene 65 2382 . + . ID=gene-RvY_00001;Name=RvY_00001-1;gbkey=Gene;gene=RvY_00001-1;gene_biotype=protein_coding;gene_synonym=RvY_00001.1;locus_tag=RvY_00001 BDGG01000001.1 DDBJ CDS 65 242 . + 0 ID=cds-GAU87103.1;Parent=gene-RvY_00001;Dbxref=NCBI_GP:GAU87103.1;Name=GAU87103.1;gbkey=CDS;gene=RvY_00001-1;locus_tag=RvY_00001;product=hypothetical protein;protein_id=GAU87103.1 BDGG01000001.1 DDBJ CDS 441 704 . + 2 ID=cds-GAU87103.1;Parent=gene-RvY_00001;Dbxref=NCBI_GP:GAU87103.1;Name=GAU87103.1;gbkey=CDS;gene=RvY_00001-1;locus_tag=RvY_00001;product=hypothetical protein;protein_id=GAU87103.1 BDGG01000001.1 DDBJ CDS 934 1359 . + 2 ID=cds-GAU87103.1;Parent=gene-RvY_00001;Dbxref=NCBI_GP:GAU87103.1;Name=GAU87103.1;gbkey=CDS;gene=RvY_00001-1;locus_tag=RvY_00001;product=hypothetical protein;protein_id=GAU87103.1 BDGG01000001.1 DDBJ CDS 1853 2382 . + 2 ID=cds-GAU87103.1;Parent=gene-RvY_00001;Dbxref=NCBI_GP:GAU87103.1;Name=GAU87103.1;gbkey=CDS;gene=RvY_00001-1;locus_tag=RvY_00001;product=hypothetical protein;protein_id=GAU87103.1
The search finds five lines of text: 4 CDS features and 1 gene feature.
What happens if we search this way?
local aloraine$ grep RvY_00001 GCA_001949185.1_Rvar.gff BDGG01000001.1 DDBJ gene 65 2382 . + . ID=gene-RvY_00001;Name=RvY_00001-1;gbkey=Gene;gene=RvY_00001-1;gene_biotype=protein_coding;gene_synonym=RvY_00001.1;locus_tag=RvY_00001 BDGG01000001.1 DDBJ CDS 65 242 . + 0 ID=cds-GAU87103.1;Parent=gene-RvY_00001;Dbxref=NCBI_GP:GAU87103.1;Name=GAU87103.1;gbkey=CDS;gene=RvY_00001-1;locus_tag=RvY_00001;product=hypothetical protein;protein_id=GAU87103.1 BDGG01000001.1 DDBJ CDS 441 704 . + 2 ID=cds-GAU87103.1;Parent=gene-RvY_00001;Dbxref=NCBI_GP:GAU87103.1;Name=GAU87103.1;gbkey=CDS;gene=RvY_00001-1;locus_tag=RvY_00001;product=hypothetical protein;protein_id=GAU87103.1 BDGG01000001.1 DDBJ CDS 934 1359 . + 2 ID=cds-GAU87103.1;Parent=gene-RvY_00001;Dbxref=NCBI_GP:GAU87103.1;Name=GAU87103.1;gbkey=CDS;gene=RvY_00001-1;locus_tag=RvY_00001;product=hypothetical protein;protein_id=GAU87103.1 BDGG01000001.1 DDBJ CDS 1853 2382 . + 2 ID=cds-GAU87103.1;Parent=gene-RvY_00001;Dbxref=NCBI_GP:GAU87103.1;Name=GAU87103.1;gbkey=CDS;gene=RvY_00001-1;locus_tag=RvY_00001;product=hypothetical protein;protein_id=GAU87103.1
The GFF file's quote lines above suggest that for some gene models, there are no exon annotations. It's possible that this file uses "CDS" and only "CDS" to indicate the locations of translated exons and maybe uses "exons" to indicate the untranslated regions (UTRs) at the 5' and 3' ends of a transcript. There are probably not very many of these because the annotation was probably done using gene-finding software, which sucks at identifying UTRs.
Let's take a look:
local aloraine$ grep exon GCA_001949185.1_Rvar.gff | head BDGG01000001.1 DDBJ exon 443104 443176 . + . ID=exon-RvY_t0001-1;Parent=rna-RvY_t0001;gbkey=tRNA;gene=tRNA-Lys(CUU);inference=ab initio prediction:Aragorn:1.2.2865%2CtRNAscan-SE:1.2366;locus_tag=RvY_t0001;product=tRNA-Lys BDGG01000001.1 DDBJ exon 468837 468908 . + . ID=exon-RvY_t0002-1;Parent=rna-RvY_t0002;gbkey=tRNA;gene=tRNA-Arg(CCG);inference=ab initio prediction:Aragorn:1.2.2865%2CtRNAscan-SE:1.2366;locus_tag=RvY_t0002;product=tRNA-Arg BDGG01000001.1 DDBJ exon 1764652 1764688 . + . ID=exon-RvY_t0003-1;Parent=rna-RvY_t0003;gbkey=tRNA;gene=tRNA-Tyr(GUA);inference=ab initio prediction:Aragorn:1.2.2865%2CtRNAscan-SE:1.2366;locus_tag=RvY_t0003;product=tRNA-Tyr BDGG01000001.1 DDBJ exon 1764702 1764737 . + . ID=exon-RvY_t0003-2;Parent=rna-RvY_t0003;gbkey=tRNA;gene=tRNA-Tyr(GUA);inference=ab initio prediction:Aragorn:1.2.2865%2CtRNAscan-SE:1.2366;locus_tag=RvY_t0003;product=tRNA-Tyr BDGG01000001.1 DDBJ exon 1908331 1908451 . - . ID=exon-RvY_r0001-1;Parent=rna-RvY_r0001;gbkey=rRNA;gene=5s_rRNA;inference=ab initio prediction:RNAmmer:1.2(score:27.1);locus_tag=RvY_r0001;product=5S ribosomal RNA BDGG01000001.1 DDBJ exon 2282898 2282979 . - . ID=exon-RvY_t0004-1;Parent=rna-RvY_t0004;gbkey=tRNA;gene=tRNA-Leu(AAG);inference=ab initio prediction:Aragorn:1.2.2865%2CtRNAscan-SE:1.2366;locus_tag=RvY_t0004;product=tRNA-Leu BDGG01000001.1 DDBJ exon 2356047 2356161 . - . ID=exon-RvY_r0002-1;Parent=rna-RvY_r0002;gbkey=rRNA;gene=5s_rRNA;inference=ab initio prediction:RNAmmer:1.2(score:82.3);locus_tag=RvY_r0002;product=5S ribosomal RNA BDGG01000001.1 DDBJ exon 2410614 2410728 . - . ID=exon-RvY_r0003-1;Parent=rna-RvY_r0003;gbkey=rRNA;gene=5s_rRNA;inference=ab initio prediction:RNAmmer:1.2(score:82.3);locus_tag=RvY_r0003;product=5S ribosomal RNA BDGG01000001.1 DDBJ exon 2418110 2418224 . - . ID=exon-RvY_r0004-1;Parent=rna-RvY_r0004;gbkey=rRNA;gene=5s_rRNA;inference=ab initio prediction:RNAmmer:1.2(score:79.3);locus_tag=RvY_r0004;product=5S ribosomal RNA BDGG01000001.1 DDBJ exon 2530911 2531025 . + . ID=exon-RvY_r0005-1;Parent=rna-RvY_r0005;gbkey=rRNA;gene=5s_rRNA;inference=ab initio prediction:RNAmmer:1.2(score:84.4);locus_tag=RvY_r0005;product=5S ribosomal RN
All of the above lines look like they are coming from an RNA-prediction program called "Aragorn". Cool cool cool.
Are there any "exons" associated with protein-coding genes?
Let's find out:
local aloraine$ grep -w exon GCA_001949185.1_Rvar.gff | grep protein local aloraine$
The preceding line finds all instances of the whole word "exon" in the target GFF file and then pipes the output to another grep command that searches for instances of the word "protein." Nothing is found. So it appears that all the protein coding genes, hypothetical or otherwise, have zero "exon" features in them.
So, to assemble gene models for protein-coding genes, we have to find all the CDS features and then assemble them into gene models in our code.
The RNA-genes will need to be assembled in a different way.
Investigating an example RNA gene:
local aloraine$ grep rna-RvY_t0002 GCA_001949185.1_Rvar.gff BDGG01000001.1 DDBJ tRNA 468837 468908 . + . ID=rna-RvY_t0002;Parent=gene-RvY_t0002;gbkey=tRNA;gene=tRNA-Arg(CCG);inference=ab initio prediction:Aragorn:1.2.2865%2CtRNAscan-SE:1.2366;locus_tag=RvY_t0002;product=tRNA-Arg BDGG01000001.1 DDBJ exon 468837 468908 . + . ID=exon-RvY_t0002-1;Parent=rna-RvY_t0002;gbkey=tRNA;gene=tRNA-Arg(CCG);inference=ab initio prediction:Aragorn:1.2.2865%2CtRNAscan-SE:1.2366;locus_tag=RvY_t0002;product=tRNA-Arg
Looks like non-coding genes are constructed from exons that get grouped using a "Parent" gene identifier in the same way that protein-coding genes do.
Examining errors and warnings emitted by gff3ToBedDetail.py:
warnings.warn("Two lines with the same gene id: %s"%gene_id)
/Users/aloraine/src/genomesource/Mapping/Parser/Gff3.py:49: UserWarning: Two lines with the same gene id: gene-RvY_00053-2
The extra feature field attribute "gene id" is getting repeated in every line. I need to look into if this is going to be a problem or not.
Investigating:
BDGG01000001.1 DDBJ gene 115830 115868 . - . ID=gene-RvY_00053-2;Name=RvY_00053;gbkey=Gene;gene=RvY_00053;gene_biotype=protein_coding;gene_synonym=RvY_00053.2;is_ordered=true;locus_tag=RvY_00053-2 BDGG01000001.1 DDBJ gene 115332 115577 . - . ID=gene-RvY_00053-2;Name=RvY_00053;gbkey=Gene;gene=RvY_00053;gene_biotype=protein_coding;gene_synonym=RvY_00053.2;is_ordered=true;locus_tag=RvY_00053-2 ... (more, not shown)
It appears the same entity with extra feature "ID=gene-RvY_00053-2" appears twice in the file, with different coordinates.
I don't think this GFF file is usable because it uses the same ID for entities with different coordinates. Looking into whether there is another option we can use instead.
Contacting NCBI for help:
Hello! I need your help with the gene model annotation data file for this genome assembly: https://www.ncbi.nlm.nih.gov/datasets/genome/GCA_001949185.1/
I am working with this annotation file: GCA_001949185.1_Rvar_4.0_genomic.gff.gz. (It is available from the "ftp" link.)
The file has a problem I hope you can help me with.
Many lines in the file contain duplicated "ID" values and nonsensical "Parent" values.
For example, there are two lines with ID value of gene-RvY_00053-2, but they have different coordinates.
My understanding of GFF is that all ID values need to be unique within a file. This is because sub-features (e.g., CDS, exon, etc) have to be assembled into coherent gene models by referencing the same Parent. If sub-features belonging to different gene models reference the same Parent identifier, then there is no way to create coherent gene models from them.
Could you help me understand the proper way to use this file?
Thank you!!!
Ann Loraine, PhD
Professor, Bioinformatics and Genomics
UNC Charlotte
Update:
- Opened 2bit file as a new "custom" genome in IGB
- Opened GFF file from Genbank in IGB, selected load model "whole genome"
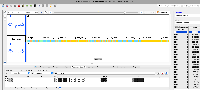
- Looked up a multi-exon gene model using NCBI's Genome Data Viewer page. Identified RvY_01679 as an example. It has two gene models and multiple exons per gene model.
- Searched using RvY_01679 as the query in IGB using Advanced Search tab. IGB found the gene but when I went to its location in IGB by double-clicking the search results, the location appeared to be empty. So, it looks like the file is getting parsed without error and some aspects of the data are getting imported, but IGB cannot draw the gene model.
- Images are attached
Note: This is an example of how seeing the same region in two different genome browsers helps science. In this case, it helps with trouble-shooting a nonsensical data file.
My assessment of what to do next:
- Wait for NCBI to get back to us.
Ann Loraine - I have added a bed file (R_varieornatus_YOKOZUNA-1_Nov_2016_ncbiGene.bed) of the R varieornatus annotation to the Google Drive: https://drive.google.com/drive/folders/1acW5w8dMGvxit3kw9i_jiW9ZOUqV6W8X?usp=drive_link
The file came from the UCSC Table Browser, see image below:
The table is listed as hub_3041413_ncbiGene, under track Gene Models
The description page for the R varieornatus gene models on the UCSC genome browser are listed as using Assembly: hub_3041413_Rvar_4.0 Nov. 2016 R.varieornatus (YOKOZUNA-1 2016 tardigrades) and
The Gene model track for the 09 Nov 2016 Ramazzottius varieornatus/GCA_001949185.1_Rvar_4.0 genome assembly is constructed from the gff file GCA_001949185.1_Rvar_4.0_genomic.gff.gz supplied with the genome assembly at the FTP location:
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/001/949/185/GCA_001949185.1_Rvar_4.0/
I'm reasonably sure that the gff file it links to is the same one we were using from NCBI, and it appears that the UCSC bed file is created from it.
The bed file has a 13th and 14th field, but the 14th field is a repeat of the gene name (i.e. it is a placeholder). The gff file we were attempting to use before has some additional information, such as a product field that we might be able to use as the 14th field.
I used the Note and Product tags from the GCA_001949185.1_Rvar.gff file to create a 14th description column. The updated bed file has been added to the Google Drive as R_varieornatus_YOKOZUNA-1_Nov_2016_ncbiGene.bed.gz. The genome.txt, 2bit file, and tabix index are also present in the Google Drive.
As noted by Dr. Loraine there are many duplicated ID values. In order to work around this, I retrieved the cds, rrna, and trna rows from the GCA_001949185.1_Rvar.gff file. Note that ID=id-RvY_02531 was an extra row in the GFF file that was hard to find and needed to be removed. I sorted by seqid, then start, then end, and then attributes. I used diff on the ID tag of the sorted gff to compare to the name value of the bed file and they were the same. I then extracted the Note and Product tags and used those to create the 14th description column. Much of this was done manually.
Next step: Create new genome and push files to SVN repo.
My apologies for the problems with the subversion server.
They are now fixed. You should now be able to commit your new files and directories to the subversion host as before.
The Ramazzottius varieornatus genome and annotation have been deployed to the subversion host: https://svn.bioviz.org/viewvc/genomes/quickload/R_varieornatus_YOKOZUNA-1_Nov_2016/
To test:
Will need to have the svn repository on local machine (I think)
- Do a svn update to pull the most recent changes
- Add the svn repository as a local quickload in IGB
- Look for the Ramazzottius varieornatus genome in IGB
- Check that hovering on the species shows "tardigrade water bear"
- Select the Ramazzottius varieornatus species and the Nov 2016 genome version
- Check that the annotation file loads by default
- Check that you can load sequence (click Load Sequence)
No issues found while conducting the test outlined above – I was able to add the R. varieornatus genome to IGB as a local quickload and confirm that the annotations and sequences load properly. This quickload is ready for next steps!
Next step is for Dr. Loraine to deploy the changes to the various Quickloads.
Changes deployed to QL at:
- RENCI host (primary quickload)
- UNC Charlotte host (backup quickload)
No issues found while conducting the test!
Closing ticket.

For the R. varieornatus tardigrade:
Paper: https://www.nature.com/articles/ncomms12808
Website: http://kumamushi.org/database.html
NCBI: https://www.ncbi.nlm.nih.gov/nuccore/BDGG00000000.1
Note: UCSC does have the R. varieornatus genome in the browser, but I am not seeing it in the Table Browser, UCSC API, or UCSC Genome Downloads.
UCSC Gene Models: https://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=2077278096_b5zQo29khGhNnPNEOOamtx0cKoXg&db=hub_2790993_GCA_001949185.1&c=BDGG01000001.1&g=hub_2790993_ncbiGene
The NCBI FTP site referenced as being used by UCSC: ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/001/949/185/GCA_001949185.1_Rvar_4.0/
I pulled the following files:
GCA_001949185.1_Rvar_4.0_genomic.gff.gz
GCA_001949185.1_Rvar_4.0_genomic.fna.gz
I was able to load the files in IGB and they look correct.
NCBI also has this page: https://www.ncbi.nlm.nih.gov/datasets/genome/GCA_001949185.1/
The Genome sequences (FASTA) and Annotation features (GFF) files are the same as those from the ftp.