Details
-
Type:
Bug
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: 10.1.0
-
Labels:None
-
Story Points:0.25
-
Epic Link:
-
Sprint:Fall 2
Description
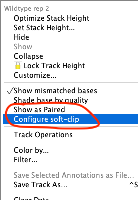
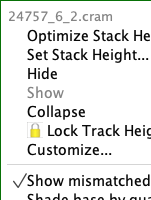
Situation: BAM/SAM files have additional right-click menu options "Show as Paired" and "Configure soft-clip" that should also be available for CRAM files, but are missing.
Task: Enable the right-click menu options listed above for CRAM files.
To reproduce the issue:
- Start IGB
- Add the smoke testing quickload as a new data provider.
- Select the human genome version H_Sapien_Dec_2013
- Add the cram and bam files (I don't think you need to load data, but see the file formats testing page for the location of where to load data in IGB if needed)
- Right-click on the track names and look for "Show as Paired" and "Configure soft-clip"
Attachments
Activity
| Field | Original Value | New Value |
|---|---|---|
| Epic Link | IGBF-1765 [ 17855 ] |
| Description |
Situation: BAM/SAM files have additional right-click menu options "Show as Paired" and "Configure soft-clip" that should also be available for CRAM files, but are missing.
Task: Enable the right-click menu options listed above for CRAM files. To reproduce the issue: Start IGB Add the [smoke testing quickload|https://quickload-testing.s3.amazonaws.com/smokeTestingQuickload/] as a new data provider. Select the human genome version H_Sapien_Dec_2013 Add the cram and bam files (I don't think you need to load data, but see the [file formats testing page|https://wiki.bioviz.org/confluence/display/ITD/File+Formats] for the location of where to load data in IGB if needed) Right-click on the track names and look for "Show as Paired" and "Configure soft-clip" |
Situation: BAM/SAM files have additional right-click menu options "Show as Paired" and "Configure soft-clip" that should also be available for CRAM files, but are missing.
Task: Enable the right-click menu options listed above for CRAM files. To reproduce the issue: # Start IGB # Add the [smoke testing quickload|https://quickload-testing.s3.amazonaws.com/smokeTestingQuickload/] as a new data provider. # Select the human genome version H_Sapien_Dec_2013 # Add the cram and bam files (I don't think you need to load data, but see the [file formats testing page|https://wiki.bioviz.org/confluence/display/ITD/File+Formats] for the location of where to load data in IGB if needed) # Right-click on the track names and look for "Show as Paired" and "Configure soft-clip" |
| Assignee | Jaya Sravani Sirigineedi [ jsirigin ] |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Attachment | CRAM_menu_options.png [ 18524 ] |
| Status | In Progress [ 3 ] | Needs 1st Level Review [ 10005 ] |
| Assignee | Jaya Sravani Sirigineedi [ jsirigin ] | Nowlan Freese [ nfreese ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Assignee | Nowlan Freese [ nfreese ] | Jaya Sravani Sirigineedi [ jsirigin ] |
| Status | First Level Review in Progress [ 10301 ] | Ready for Pull Request [ 10304 ] |
| Status | Ready for Pull Request [ 10304 ] | Pull Request Submitted [ 10101 ] |
| Assignee | Jaya Sravani Sirigineedi [ jsirigin ] |
| Assignee | Ann Loraine [ aloraine ] |
| Status | Pull Request Submitted [ 10101 ] | Reviewing Pull Request [ 10303 ] |
| Story Points | 2 | 0.25 |
| Status | Reviewing Pull Request [ 10303 ] | Merged Needs Testing [ 10002 ] |
| Assignee | Ann Loraine [ aloraine ] |
| Status | Merged Needs Testing [ 10002 ] | Post-merge Testing In Progress [ 10003 ] |
| Fix Version/s | 10.1.0 [ 11000 ] |
| Assignee | Jaya Sravani Sirigineedi [ jsirigin ] |
| Resolution | Done [ 10000 ] | |
| Status | Post-merge Testing In Progress [ 10003 ] | Closed [ 6 ] |