Details
-
Type:
Task
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: 10.1.0
-
Labels:None
-
Story Points:3
-
Epic Link:
-
Sprint:Fall 2
Description
Situation: UCSC REST data is currently organized by file "type", which often separates data into large folders of bed, genePred, etc. The UCSC genome browser website uses groupings based on the data itself, for example, Genes and Gene Predictions, Expression and Regulation, etc.
Task: Organize the UCSC REST data by the "group" value provided through the API.
Attachments
Issue Links
- relates to
-
IGBF-3689 Make folders inside the UCSC Rest folder
-
- Closed
-
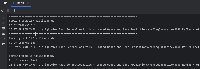
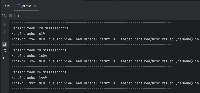
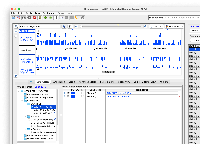
Mapping the "group" values to what UCSC shows in their browser.
Note: I looked at the T2T, the hg38, and mm39 API responses. The below table should encompass most of the "group" values that are present at UCSC, though I haven't been able to find a webpage or API call that lists all of the mappings.
Unfortunately where this gets complicated is for the data that are in folders on UCSC site. Since we are passing the trackLeavesOnly=1 parameter to the API, we aren't able to see the "parent". I've included an example below where I do not pass the trackLeavesOnly=1 to the API.
"T2T_Encode": { "compositeContainer": "TRUE", "shortLabel": "T2T Encode", "type": "bed 3", "longLabel": "T2T Encode Reanalysis", "sortOrder": "cell=+ ab=+ view=+", "subGroup1": "cell Cell_Line CL_22Rv1=22Rv1 BE2C=BE2C C4-2B=C4-2B Caco-2=Caco-2 HAP-1=HAP-1 HL-60=HL-60 MG63=MG63 RWPE1=RWPE1 RWPE2=RWPE2 SJCRH30=SJCRH30 SJSA1=SJSA1 VCaP=VCaP bmec=brain_microvascular_endothelial_cell ecopm=epithelial_cell_of_prostate_male", "visibility": "hide", "subGroup2": "ab Antibody CTCF=CTCF Control=Control H3K27ac=H3K27ac H3K27me3=H3K27me3 H3K36me3=H3K36me3 H3K4me1=H3K4me1 H3K4me3=H3K4me3 H3K9me3=H3K9me3", "subGroup3": "view View coverage=Coverage enrichment=Enrichment peaks=Macs2_Peaks LOpeaks=Macs2_hg38_LO_Peaks", "group": "regulation", "html": "T2T_Encode.html", "compositeTrack": "on", "dimensions": "dimX=cell dimY=ab", "T2T_Encode_Coverage": { "compositeViewContainer": "TRUE", "shortLabel": "Encode Coverage", "type": "bigWig", "longLabel": "Encode Coverage", "parent": "T2T_Encode", "visibility": "full", "maxHeightPixels": "100:20:10", "windowingFunction": "mean", "view": "coverage", "autoScale": "on", "T2T_Encode_Coverage_BE2C.Control": { "shortLabel": "BE2C Control", "type": "bigWig", "longLabel": "BE2C Control Coverage", "parent": "T2T_Encode_Coverage", "parentParent": "T2T_Encode", "subGroups": "cell=BE2C ab=Control view=coverage", "bigDataUrl": "/gbdb/hs1/encode/coverage/BE2C.Control.chm13v2.0.bw" },The parent, T2T_Encode, has a group value of regulation. It is a folder that contains subfolders (for example T2T_Encode_Coverage) which then contain data (for example T2T_Encode_Coverage_BE2C.Control). I think the best approach would be to emulate the organization that UCSC has, that way a user would see the same organization.