Details
-
Type:
 Support
Support
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Labels:None
Description
From: KA Atkins <kelly.atkins@bristol.ac.uk>
Reply-To: "kelly.atkins@bristol.ac.uk" <kelly.atkins@bristol.ac.uk>
Date: Tuesday, April 21, 2015 at 7:33 AM
To: "Norris, David" <dcnorris@uncc.edu>, "Loraine, Ann" <Ann.Loraine@uncc.edu>
Subject: Question regarding Depth Graph feature in IGB
Hi,
I am wondering if you can help me. I am a new user to IGB and am finding the programme very useful in visualising my recent sequencing data. I have a rather menial question but cannot find the answer in the literature.
Would you be able to tell me what the difference is between Depth Graph (All) and Depth Graph (Start) located under the Track Operation function.
Many thanks for your time,
Kelly
–
Kelly Atkins
PhD Student
School of Biological Sciences, Office 308
Bristol Life Sciences Building
University of Bristol,
24 Tyndall Ave
Bristol BS8 1TQ.
UK
tel +44 (0)117 394 1448
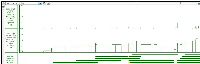
I'm not sure if this is documented anywhere in the Users Guide. If not, we should do that.
I think also that a nice explanation is located in the BioViz News page from when we added this new feature to a release.