Details
-
Type:
 Updated Genome
Updated Genome
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: None
-
Labels:
-
Story Points:0.5
-
Epic Link:
Description
Mason replied to user inquiry:
Thank you for taking the time to contact us with your question. You asked why the default RefGene track in IGB does not display all of the gene variants seen in the UCSC refgene track for the mm9 genome.
________
I opened the same two tracks (I think) in the M_musculus_Dec_2011 genome.
Black is the default track that IGB opens with this genome.
Blue is the refGene track in the UCSC folder.
Looking at the Fn1 gene, the two tracks appear to the the same, Which is very different from the image Mason attached. Mason, did I open the same files as you did?
But some of the other genes from your image (Ankar and Igfbp2) do appear to be different between the two tracks, just like your image shows.
I assume the differences between the tracks are due to differences in where the data came from. Perhaps they are from different versions of the annotations. But both having the same name and no distinguishing info would imply that they are the same.
I can look a the quickload more to see if there are notes about how each of these was made and where the data came from. But I will need to dig a bit.
_________
Thanks for looking into this Ivory!
The difference may be that the data in Quickload are out of date compared to what's in the UCSC genome browser. To investigate, you can retrieve the data from UCSC using the Table Browser and compare to the refseq data file in Quickload.
We can easily replace the data in quickload - just need to update the documentation for it.
Check the Developer's guide for details - review the information about updating a genome version.
If I'm correct, the "action item" for this Jira issue would be to update the data file for mouse and post it on Quickload, using svn to commit it.
-Ann
Attachments
Issue Links
- relates to
-
HELP-268 All RefGene variants not displayed
- Closed
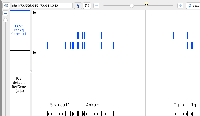
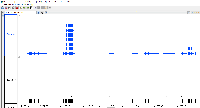
Hello again Ann and Ivory,
I continued looking into this by downloading the mm9 genome using the UCSC Table Browser, as Ann recommended (see screenshot). It appears to me that Ann is correct in that we should update the mm9 genome (and possibly others) in IGB Quickload. I see that Ivory has already created an issue for this; thanks for doing that Ivory. The other thing that is very concerning to me though is that Ivory's screenshot shows 7 transcripts for the Fn1 gene from IGB Quickload, but my screenshot only shows 1 transcript. How can this be if it is using the same data file? I tested on Windows and Mac just to make sure, but I am still seeing only 1 transcript fore the Fn1 gene. I think we should dig into this a little more to see why this is happening. Does anybody have any thoughts on this?
Thanks,
Mason