Details
-
Type:
Task
-
Status: Closed (View Workflow)
-
Priority:
Minor
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: None
-
Labels:
-
Story Points:1
-
Sprint:Winter 2018 Sprint 3, Spring 2019 Sprint 1
Description
If you open a file in IGB without selecting a genome, you can view the file but there is no genome sequence.
So the Load Sequence option probably shouldn't be enabled.
As it is, you can click the Load Sequence button, and you get an error that says:
"Couldn't locate the partial sequence. Try loading the full sequence."
This error message does not make sense given the context. (In other contexts it does make sense.)
The error message for this particular workflow should be more informative. Or, the button "Load Sequence" should not be active when no sequence file is available.
The task:
- Investigate and identify files/code involved.
- Determine: is there any way for IGB to determine if the reference genome sequence is available to load before showing that message?
- Design a solution that improves user experience.
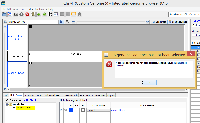
Test file - download and load without 1st selecting a genome: