Details
-
Type:
Improvement
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: 9.0.1 Minor Release
-
Labels:None
-
Story Points:2
-
Epic Link:
-
Sprint:Fall 2017
Description
Combining two graph tracks is really ideal for visualizing the chip seq data with the IP sample against the input sample as a backdrop. Once I've joined two tacks in IGB, the new track has the name "Joined Graphs". There does not seem to be any way to change the track name shown for the new joined track. Since I have a joined track for each sample, it is important that I be able to distinguish them in figures.
Thanks
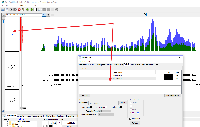
This looks like a good place for Ivory to get started with IGB code.