Details
-
Type:
Task
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: None
-
Labels:None
-
Story Points:0.5
-
Epic Link:
-
Sprint:Spring 8 : 24 Apr to 8 May, Spring 8 : 11 May to 25 May, Spring 9 : 25 May to 8 Jun
Description
BioViz Connect has a feature that lets users run analysis algorithms on files using CyVerse cloud resources. This is super useful because often people want to see output of visual analytics functions that are too compute or data intensive to run on a personal computer.
So we (IGB CyVerse core team) have created and published analysis "apps" within CyVerse itself, thus making them available for users to run and then view the output in IGB.
Recently we published an App for generating scaled coverage graphs. This uses a tool from the DeepTools suite.
Currently, the documentation for the App within BioViz Connect contains a weird-looking string - %INPUT%bam. (Please see attached screen capture.) It is not clear what this means and so this could be a bit confusing for users.
For this task, investigate why this is being shown. If feasible, propose a way to remove it, unless it is trying to convey some kind of useful information, in which case we should probably try to think of better wording.
Note: We might leave it – depends on the level of difficulty involved! Thus the goal of this task is to "investigate" only.
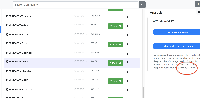
cc: Nowlan Freese to keep you in the loop.