Details
-
Type:
Task
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: None
-
Labels:None
-
Story Points:1
-
Epic Link:
-
Sprint:Spring 8 : 11 May to 25 May, Spring 9 : 25 May to 8 Jun
Description
Situation: When a user clicks on View in IGB, BioViz Connect checks to see if IGB is currently running. If IGB is not running, a toast stating "Error, IGB not running" appears. This is somewhat confusing to users as it does not lead the user to the IGB downloads page or provide additional information for how to get IGB running.
Task: Instead of the toast, use a popup/alert/message (similar to the make file public alert) when a user clicks View in IGB and IGB is not running. The notification should inform the user that IGB must be running to view the data, if they have not installed IGB they can download it from Bioviz. Include an OK button that closes out the notification and a button that links to https://www.bioviz.org/download.html in case they need to download IGB. This should make it more clear to the user what they need to view data in IGB and provide them a direct route to download IGB.
Attachments
Issue Links
- relates to
-
IGBF-2386 Modify IGB-is-not-running popup
-
- Closed
-
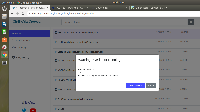
Let's use the same type of thing seen on the Genome Dashboard. If you click a genome picture, a modal dialog appears.
Please have the link open a new page and show the bioviz.org IGB Downloads page.
To see what I mean, visit:
and click an image.
Also, please note that BioViz Connect should use a better-worded message that is a bit more user-friendly than "IGB Service not running" (currently shown in Genome Dashboard.)