[~aloraine]
My recommendation would be to revert these changes.
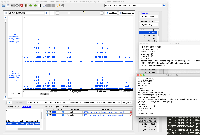
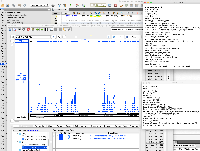
The changes in the PR above do not appear to fix the issue. Testing on 9.1.6 on the attached fimo.gff file loads the data exactly the same as 9.1.4. The main issue is that only a small number of the data appear in IGB compared to examination of the file itself. However, the data that do appear have correct formatting.
To force IGB to show all of the data, the ##gff-version 3 first line comment can be removed from the fimo.gff file. This appears to cause IGB to parse the data not as a GFF3 file, but as a GTF file.
Note that GTF files parse the attribute column as: attribute_name “attribute_value”; attribute_name “attribute_value”;
whereas GFF3 files parse the attribute column as: ID=cds00004;Parent=mRNA00001,mRNA00002;Name=edenprotein.4
The attribute values in the fimo.gff file are formatted according to the GFF3 spec as: Name=NNNNKCMTYGRDWHYYGTG_1-;Alias=MEME-2;
So by removing ##gff-version 3 and loading the fimo.gff file as a GTF through IGB the attribute column values appear with the equals signs. By stripping the equals signs in the parsing this causes the fimo.gff file to appear to be loaded correctly (through the GTF parser), but it is affecting the parsing of GTF files.
In 9.1.6 the GTF_HomoSapienChr1Only.gtf file loads all of the data in a single row and the attribute column does not contain the correct values. In 9.1.4, the data appear correct visually, though the attribute column also does not appear to be parsed correctly.
After reading through this description of GTF/GFF file formats I found this sentence describing GFF3 ID: "The ID attribute is required for features that have children (e.g. gene and mRNAs), or for those that span multiple lines, but are optional for other features." The ID field is optional and the NAME field does not have to be unique: "Unlike IDs, there is no requirement that the Name be unique within the file."
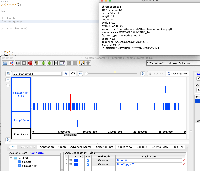
I think it may be worth trying to understand further why the fimo.gff file is not displaying correctly in IGB. Edit: The fimo.gff file includes a non-unique ID attribute, which causes incorrect parsing of the file. Removal of the ID attribute from the fimo file appears to fix the issue.
Task
Major
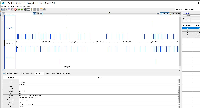
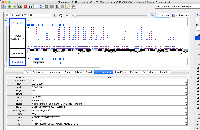
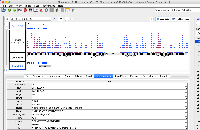
The file does not pass this gff3 validator: http://genometools.org/cgi-bin/gff3validator.cgi
This file seems odd to me. Would need to do a comparison with the gff3 specifications to double-check the file formatting and compare that to the gff3 expectations of IGB.