Details
-
Type:
Task
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: None
-
Labels:None
-
Story Points:0.5
-
Epic Link:
-
Sprint:Spring 8 : 11 May to 25 May, Spring 9 : 25 May to 8 Jun
Description
Dr. Beth Krizek (krizek@biol.sc.edu) has let us know about a problem with parsing the attached file.
It is GFF format, but when we view it in IGB, only a small part of the data are being shown.
When you click on one of the items, the "Id" field does not exactly match the "Id" fields in the GFF "extra feature" field, the section of semi-colon-separated characters in the final field.
Please investigate why this is happening. Is there a problem with our GFF parsing code, or is it an issue with the formatting of this file?
In addition, take a look at the unit testing code for this format. If the data in this file are OK, we should use some lines of data in the file to add new test cases to the testing code.
The file was generated using a program called "FIMO" which is a popular tool in bioinformatics. Therefore we should do our best to support visualizing the output in IGB.
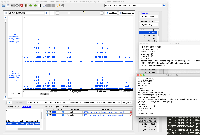
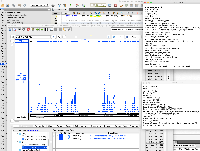
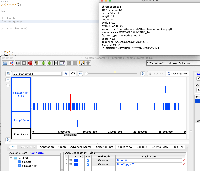
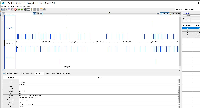
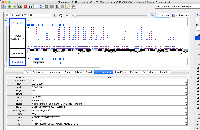
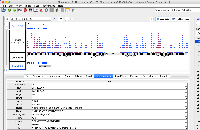
The file does not pass this gff3 validator: http://genometools.org/cgi-bin/gff3validator.cgi
This file seems odd to me. Would need to do a comparison with the gff3 specifications to double-check the file formatting and compare that to the gff3 expectations of IGB.