Details
-
Type:
New Feature
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: None
-
Labels:None
-
Story Points:0.5
-
Epic Link:
-
Sprint:Spring 8 : 11 May to 25 May, Spring 9 : 25 May to 8 Jun, Summer 1: 8 Jun - 19 Jun
Description
Note: Do this after IGBF-2386 has been merged into master.
The new genome dashboard application (https://www.bioviz.org/genome-dashboard) shows images representing different species for which genome sequence data are available.
When users click an image, the latest genome version for the featured species is supposed to load into IGB.
However, this only works if IGB is currently running.
So the Genome Dashboard App has some code that checks to see if IGB is running.
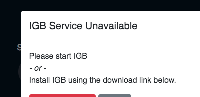
If IGB is not running, then it shows a popup to let them know to start IGB.
The popup includes button labeled "Download Now" which takes the user to the IGB download page.
This is great, but it would be more convenient for users if the button actually were to trigger downloading of the installer for the user's platform. This would save the user a bunch of clicks and would also get them started using IGB faster.
For this task, modify the button code so that matches what happens when users visit the IGB downloads page (https://www.bioviz.org/download.html), which figures out the user's platform and then dynamically builds a link to the installer they need.
Note that some code for this already exists on the BioViz.org Web site and in this repository: https://bitbucket.org/lorainelab/bioviz
Also note that genome-dashboard, although in a different repository from the bioviz code, is meant to be deployed onto the same host as the bioviz Web site code.
Therefore you should re-use and/or re-design existing BioViz javascript code for this task. Otherwise, there will be two separate implementations of the same functionality in our code bases, which makes maintenance unnecessarily complex. The possible need for re-factoring is why the story points are so high.
Sameer is doing an internship this summer, but I think it would be fine to consult with him on this as needed.
Note: Please do IGBF-2386 first.
cc: Sameer Shanbhag
Javascript that supports this functionality is defined on
Javascript to refer to
<script> $(window).on("load",function (e) { var parser = new UAParser(); var os = parser.getOS().name; var systemConfig = parser.getCPU(); if (os == "Linux" || os == "CentOS" || os == "Fedora" || os == "FreeBSD" || os == "Debian" || os == "DragonFly" || os == "RedHat" || os == "Mint" || os == "GNU" || os == "Ubuntu") { $("#linuxDownload").removeClass("hide"); } if (os == "Windows [Phone/Mobile]" || os == "Windows") { if (systemConfig.architecture == "amd64" || systemConfig.architecture == "ia64" || systemConfig.architecture == "arm64" || systemConfig.architecture == "irix64" || systemConfig.architecture == "mips64" || systemConfig.architecture == "sparc64") { $("#windows64Download").removeClass("hide"); } else { $("#windows64Download").removeClass("hide"); $("#windows32Download").removeClass("hide"); } $("#windows_warning").removeClass("hide"); } if (os == "Mac OS X" || os == "Mac OS") { $("#osxDownload").removeClass("hide"); } }); </script>Task:
c.c. Prof. [~aloraine]