Details
-
Type:
Task
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: None
-
Labels:None
-
Story Points:3
-
Epic Link:
-
Sprint:Summer 5: 3 Aug - 14 Aug, Summer 6: 17 Aug - 28 Aug, Summer 7: 31 Aug - 11 Sep, Fall 1: 14 Sep - 25 Sep
Description
Welcome to your first on-boarding task for the IGB core development team!
We are super excited you have joined us! We hope you will enjoy your time with us and will make big contributions to the code base and the IGB community.
For this on-boarding task, please do the following:
- Go to https://canvas.instructure.com and make a free account.
- Log in and enroll in https://canvas.instructure.com/courses/1164217
- In Module I of the course, read: IGB Background and Intro to IGB Data Visualization
- Download and install the current release of IGB onto your computer.
- Using IGB, copy the actions shown in this IGB video: https://www.youtube.com/watch?v=LjPNT1k9cqk
- Re-create the scene shown at the very end of the video.
- Use the camera button in the IGB toolbar to take a picture of IGB showing the scene at the end of the video; upload it here
Reviewers:
- Watch https://www.youtube.com/watch?v=LjPNT1k9cqk
- Confirm that the assignee has uploaded an image with graphs loaded and configured to the same scale as each other, as shown in the video
- Confirm that the assignee has enrolled in the class.
Attachments
Issue Links
- blocks
-
IGBF-1763 On-boarding: Step 1 - set up your account in bitbucket and jira
-
- Closed
-

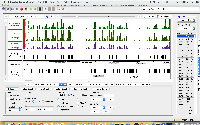
Use "attach files" option under More menu in Jira ticket. This is better than embedding the image into the page because the image is too big.