Prototype 1
This idea is based on the output from CRISPRon.
To view the prototype-1.bed file in IGB:
Open the human genome in IGB (H_sapiens_Dec_2013)
Navigate to chr17:43,125,265-43,125,369
Load the prototype-1.bed file and click Load Data
Right-click on the track label for prototype-1.bed and select Color by...
In the Color By window, select RGB in the dropdown menu and click OK
Pros: The bed file includes multiple pieces of data in the Name column, which decreases the size of the file compared to using bedDetail while still including necessary information such as the CRISPR target id, sequence, and PAM site. The RGB value could equate to the score for that particular CRISPR target, and thus using IGB's Color by... functionality allows for fast visual analysis of which CRISPR targets would work best within a particular location.
Cons: Using bedDetail and placing the CRISPR target id in the 13th column and the sequence of the target/PAM site in the 14th column may be more intuitive. Alternatively, only including the six columns (up to strand) would further decrease the file size while still enabling visual analysis by color using Color by... > Score and editing the heatmap (though this requires additional steps for the user, including knowing to set the range of the heatmap to appropriate values.
CRISPRon uses IGV JavaScript to display the results on their webpage. The results can be exported as a bed file. The only difference between the prototype-1.bed file and the CRISPRon output is that the thickStart and thickEnd columns are set to chromStart in the prototype-1.bed file. The initial CRISPRon output had the thickStart match the chromStart and the thickEnd match the chromEnd. This resulted in IGB showing the predicted amino acids as part of the annotation, which I found to distract from the visualization of CRISPR target sites.
Task
Major
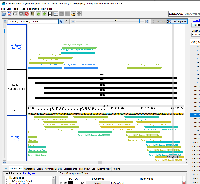
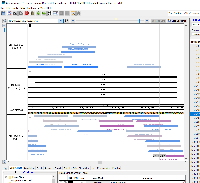
Prototype 1
This idea is based on the output from CRISPRon.
To view the prototype-1.bed file in IGB:
Open the human genome in IGB (H_sapiens_Dec_2013)
Navigate to chr17:43,125,265-43,125,369
Load the prototype-1.bed file and click Load Data
Right-click on the track label for prototype-1.bed and select Color by...
In the Color By window, select RGB in the dropdown menu and click OK
Pros: The bed file includes multiple pieces of data in the Name column, which decreases the size of the file compared to using bedDetail while still including necessary information such as the CRISPR target id, sequence, and PAM site. The RGB value could equate to the score for that particular CRISPR target, and thus using IGB's Color by... functionality allows for fast visual analysis of which CRISPR targets would work best within a particular location.
Cons: Using bedDetail and placing the CRISPR target id in the 13th column and the sequence of the target/PAM site in the 14th column may be more intuitive. Alternatively, only including the six columns (up to strand) would further decrease the file size while still enabling visual analysis by color using Color by... > Score and editing the heatmap (though this requires additional steps for the user, including knowing to set the range of the heatmap to appropriate values.
CRISPRon uses IGV JavaScript to display the results on their webpage. The results can be exported as a bed file. The only difference between the prototype-1.bed file and the CRISPRon output is that the thickStart and thickEnd columns are set to chromStart in the prototype-1.bed file. The initial CRISPRon output had the thickStart match the chromStart and the thickEnd match the chromEnd. This resulted in IGB showing the predicted amino acids as part of the annotation, which I found to distract from the visualization of CRISPR target sites.