UCSC Genome Browser:
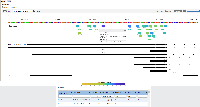
For the human and mouse genomes (tested) there is a track labelled CRISPR targets. Additional information about the track can be found here. "CRISPR target sites were annotated with predicted specificity (off-target effects) and predicted efficiency (on-target cleavage) by various algorithms through the tool CRISPOR."
The CRISPR targets are color-coded based on specificity (gray indicates CRISPR site would likely have off-target effects) or blue/red/yellow/green indicating low to high predicted efficiencies.
Mousing over the target site shows predicted specificity and efficiency scores.
Clicking on the target site opens a new web page with more detailed information, such as the guide sequence, PAM site, and specificity/efficiency scores.
Note that some of the CRISPR design sites (such as CHOPCHOP) also include links to UCSC where the predicted CRISPR sites are included as new tracks.
Ensembl genome browser:
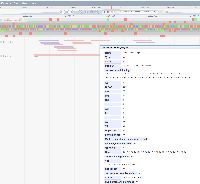
Ensembl has a track for predicted CRISPR sites for the human and mouse genomes. "NGG CRISPR sites identified by the Wellcome Trust Sanger Institute Genome Editing (WGE) resource https://doi.org/10.1093/bioinformatics/btv308, with percent in-frame and favoured mutations as predicted by FORECasT https://doi.org/10.1038/nbt.4317." Clicking on the predicted CRISPR sites opens a popup with additional information such as the predicted number of off targets. PAM sites (NGG) are marked as solid green while the rest of the predicted site is outlined in green.
To enable the CRISPR track:
Click on "Configure this page"
Select "Genome targeting"
Select "CRISPR SpCas9"
The Breaking Cas CRISPR designer uses the annotations and genomes from Ensembl, so has a wide variety of genomes available. When viewing the output of Breaking Cas there is a link to Ensembl, however, this takes the user to Ensembl but does not create a custom track with the CRISPR output.
Jbrowse:
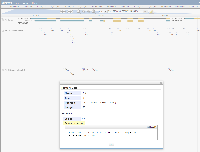
The CRISPR-PLANT designer has built-in support for JBrowse CRISPR visualization. "Different classes of gRNA spacers are loaded as individual tracks in JBrowse." Clicking on a gRNA spacer opens a popup with additional information, such as the Name, Score, Position, Length, and sequence.
The CRISPR-DT browser also uses JBrowse to show CRISPR targets. "Users can click the JBrowse link to visualize each target site in the background of genome and transcript features." The CRISPR targets are added as a separate track labelled Target site. Clicking on a CRISPR target opens a popup with additional information (looks like it is formatted as BAM/SAM).
IGV java version:
There are no details about CRISPR specific settings or tools for IGV according to their user guide or their help page.
The CRISPR design site CRISPR-SURF's output can be exported for visualization in IGV. The export is an IGV session folder with bed and bedgraph files containing scores and significant regions.
IGV javascript version:
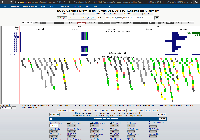
The CRISPRon CRISPR designer output can be viewed in IGV JavaScript. "Below you see the targets ranked according to their predicted cleavage efficiency. Cutoffs for selection of best targets of >= 50% efficiency and <= 20% repeat masking are used." The CRISPR sites are viewed as their own track (Best targets) and are color coded. Clicking on a CRISPR target opens a popup with the Score, Name, Target, PAM, and Masked information. The output can be exported as a bed file. Note that the user does not have to download and then upload the output to IGV JavaScript. It appears that CRISPRon has their own implementation/hosting of IGV JavaScript and pulls in the CRISPR results as a new track.
Task
Major


UCSC Genome Browser:
For the human and mouse genomes (tested) there is a track labelled CRISPR targets. Additional information about the track can be found here. "CRISPR target sites were annotated with predicted specificity (off-target effects) and predicted efficiency (on-target cleavage) by various algorithms through the tool CRISPOR."
The CRISPR targets are color-coded based on specificity (gray indicates CRISPR site would likely have off-target effects) or blue/red/yellow/green indicating low to high predicted efficiencies.
Mousing over the target site shows predicted specificity and efficiency scores.
Clicking on the target site opens a new web page with more detailed information, such as the guide sequence, PAM site, and specificity/efficiency scores.
Note that some of the CRISPR design sites (such as CHOPCHOP) also include links to UCSC where the predicted CRISPR sites are included as new tracks.
Ensembl genome browser:
Ensembl has a track for predicted CRISPR sites for the human and mouse genomes. "NGG CRISPR sites identified by the Wellcome Trust Sanger Institute Genome Editing (WGE) resource https://doi.org/10.1093/bioinformatics/btv308, with percent in-frame and favoured mutations as predicted by FORECasT https://doi.org/10.1038/nbt.4317." Clicking on the predicted CRISPR sites opens a popup with additional information such as the predicted number of off targets. PAM sites (NGG) are marked as solid green while the rest of the predicted site is outlined in green.
To enable the CRISPR track:
Click on "Configure this page"
Select "Genome targeting"
Select "CRISPR SpCas9"
The Breaking Cas CRISPR designer uses the annotations and genomes from Ensembl, so has a wide variety of genomes available. When viewing the output of Breaking Cas there is a link to Ensembl, however, this takes the user to Ensembl but does not create a custom track with the CRISPR output.
Jbrowse:
The CRISPR-PLANT designer has built-in support for JBrowse CRISPR visualization. "Different classes of gRNA spacers are loaded as individual tracks in JBrowse." Clicking on a gRNA spacer opens a popup with additional information, such as the Name, Score, Position, Length, and sequence.
The CRISPR-DT browser also uses JBrowse to show CRISPR targets. "Users can click the JBrowse link to visualize each target site in the background of genome and transcript features." The CRISPR targets are added as a separate track labelled Target site. Clicking on a CRISPR target opens a popup with additional information (looks like it is formatted as BAM/SAM).
IGV java version:
There are no details about CRISPR specific settings or tools for IGV according to their user guide or their help page.
The CRISPR design site CRISPR-SURF's output can be exported for visualization in IGV. The export is an IGV session folder with bed and bedgraph files containing scores and significant regions.
IGV javascript version:
The CRISPRon CRISPR designer output can be viewed in IGV JavaScript. "Below you see the targets ranked according to their predicted cleavage efficiency. Cutoffs for selection of best targets of >= 50% efficiency and <= 20% repeat masking are used." The CRISPR sites are viewed as their own track (Best targets) and are color coded. Clicking on a CRISPR target opens a popup with the Score, Name, Target, PAM, and Masked information. The output can be exported as a bed file. Note that the user does not have to download and then upload the output to IGV JavaScript. It appears that CRISPRon has their own implementation/hosting of IGV JavaScript and pulls in the CRISPR results as a new track.