Details
-
Type:
Task
-
Status: Closed (View Workflow)
-
Priority:
Minor
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: None
-
Labels:
-
Story Points:2
-
Epic Link:
-
Sprint:Summer 1
Description
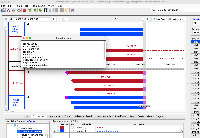
Situation: psl files (EST and mRNA) have some kind of off by one error. The listed end position is +1 compared to where the annotations actually end (positive strand alignments).
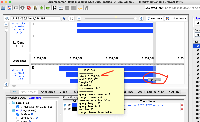
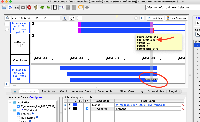
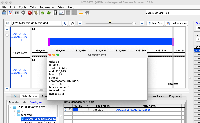
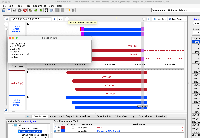
For example, if a mRNA on the positive strand says that it starts at 100 and ends at 150, in IGB the annotation will be drawn overlapping bases 100 - 149. If on the negative strand the IGB reported start is +1 to where the annotation is actually drawn. This indicates that the tEnd (https://genome.ucsc.edu/FAQ/FAQformat.html#format2) column is effectively +1 compared to what is actually drawn in IGB. Note that the annotations in IGB appear to be drawn in the correct location, i.e. intron exon boundaries line up correctly. The only issue is that the IGB reported start/end is off by one, depending on positive/negative strand.
Task: Investigate why this issue is occurring. If the issue can be fixed update this ticket.
Attachments
Issue Links
- relates to
-
IGBF-3330 Add mm39 (GRCm39) mouse genome to IGB
-
- Closed
-
Activity
| Field | Original Value | New Value |
|---|---|---|
| Epic Link | IGBF-1765 [ 17855 ] |
| Description |
Situation:
Task: |
Situation: psl files (EST and mRNA) have some kind of off by one error. The listed end position is +1 compared to where the annotations actually end (positive strand alignments).
For example, if a mRNA on the positive strand says that it starts at 100 and ends at 150, in IGB the annotation will be drawn overlapping bases 100 - 149. If on the negative strand the IGB reported start is +1 to where the annotation is actually drawn. This indicates that the tEnd (https://genome.ucsc.edu/FAQ/FAQformat.html#format2) column is effectively +1 compared to what is actually drawn in IGB. Note that the annotations in IGB appear to be drawn in the correct location, i.e. intron exon boundaries line up correctly. The only issue is that the IGB reported start/end is off by one, depending on positive/negative strand. This issue is present in older psl files (M_musculus_Dec_2011) and on older versions of IGB (tested 9.0.2). Task: Investigate why this issue is occurring. If the issue can be fixed update this ticket. |
| Description |
Situation: psl files (EST and mRNA) have some kind of off by one error. The listed end position is +1 compared to where the annotations actually end (positive strand alignments).
For example, if a mRNA on the positive strand says that it starts at 100 and ends at 150, in IGB the annotation will be drawn overlapping bases 100 - 149. If on the negative strand the IGB reported start is +1 to where the annotation is actually drawn. This indicates that the tEnd (https://genome.ucsc.edu/FAQ/FAQformat.html#format2) column is effectively +1 compared to what is actually drawn in IGB. Note that the annotations in IGB appear to be drawn in the correct location, i.e. intron exon boundaries line up correctly. The only issue is that the IGB reported start/end is off by one, depending on positive/negative strand. This issue is present in older psl files (M_musculus_Dec_2011) and on older versions of IGB (tested 9.0.2). Task: Investigate why this issue is occurring. If the issue can be fixed update this ticket. |
*Situation*: psl files (EST and mRNA) have some kind of off by one error. The listed end position is +1 compared to where the annotations actually end (positive strand alignments).
For example, if a mRNA on the positive strand says that it starts at 100 and ends at 150, in IGB the annotation will be drawn overlapping bases 100 - 149. If on the negative strand the IGB reported start is +1 to where the annotation is actually drawn. This indicates that the tEnd (https://genome.ucsc.edu/FAQ/FAQformat.html#format2) column is effectively +1 compared to what is actually drawn in IGB. Note that the annotations in IGB appear to be drawn in the correct location, i.e. intron exon boundaries line up correctly. The only issue is that the IGB reported start/end is off by one, depending on positive/negative strand. *Task*: Investigate why this issue is occurring. If the issue can be fixed update this ticket. |
| Attachment | M_musculus_Jun_2020_TEST_mrna.psl [ 17894 ] |
| Attachment | negative.png [ 17895 ] | |
| Attachment | positive.png [ 17896 ] |
| Attachment | negative.png [ 17895 ] |
| Attachment | positive.png [ 17896 ] |
| Attachment | negative.png [ 17897 ] | |
| Attachment | positive.png [ 17898 ] |
| Labels | intermediate |
| Sprint | Summer 4 2023 June 26 [ 173 ] |
| Sprint | Summer 1 [ 195 ] |
| Sprint | Summer 1 [ 195 ] | Spring 10 [ 194 ] |
| Rank | Ranked lower |
| Sprint | Spring 10 [ 194 ] |
| Sprint | Summer 1 [ 195 ] |
| Rank | Ranked higher |
| Assignee | Jaya Sravani Sirigineedi [ jsirigin ] |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Attachment | Screenshot 2024-06-04 at 4.07.55 PM.png [ 18407 ] |
| Attachment | Screenshot 2024-06-04 at 3.51.29 PM.png [ 18408 ] |
| Attachment | Screenshot 2024-06-04 at 3.54.27 PM.png [ 18409 ] |
| Attachment | Screenshot 2024-06-04 at 3.54.10 PM.png [ 18410 ] |
| Status | In Progress [ 3 ] | To-Do [ 10305 ] |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Status | In Progress [ 3 ] | Needs 1st Level Review [ 10005 ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Status | First Level Review in Progress [ 10301 ] | Needs 1st Level Review [ 10005 ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Status | First Level Review in Progress [ 10301 ] | Ready for Pull Request [ 10304 ] |
| Status | Ready for Pull Request [ 10304 ] | Pull Request Submitted [ 10101 ] |
| Status | Pull Request Submitted [ 10101 ] | Reviewing Pull Request [ 10303 ] |
| Status | Reviewing Pull Request [ 10303 ] | Merged Needs Testing [ 10002 ] |
| Status | Merged Needs Testing [ 10002 ] | Post-merge Testing In Progress [ 10003 ] |
| Resolution | Done [ 10000 ] | |
| Status | Post-merge Testing In Progress [ 10003 ] | Closed [ 6 ] |