Details
-
Type:
Task
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: None
-
Labels:None
-
Story Points:2
-
Epic Link:
-
Sprint:Summer 5 2023 July 10
Description
I attempted to run a search of InterPro using ProtAnnot but the search did not execute properly. (See attached image for a view of ProtAnnot.)
It looks like maybe the InterProScan Web service API has changed and ProtAnnot needs to be updated.
Note:
To understand how to run a search of InterPro using ProtAnnot, watch the video linked from the top page of https://apps.bioviz.org. The first part of the video explains how to install an IGB App using the App Store. You can skip the first part, since you probably should work with the development (current master branch) version of ProtAnnot available from the Downloads section of a ProtAnnot bitbucket repository. To just see how to run a search with ProtAnnot, you can skip ahead to 1:43 minutes into the video.
The error, copied from IGB "log" tab:
01:20:10.863 INFO com.affymetrix.igb.IGB - Completed loading sequence for Chr1 : 7,284,963 - 7,290,069 from IGB Quickload http://lorainelab-quickload.scidas.org/quickload/
01:21:04.688 INFO o.l.i.p.i.InterProscanServiceRest - iprscan5-R20230710-072102-0693-74435734-p1m
01:21:05.129 ERROR o.l.i.p.i.InterProscanServiceRest - No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED
java.lang.IllegalArgumentException: No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED
at java.lang.Enum.valueOf(Enum.java:238) ~[na:1.8.0_332]
at org.lorainelab.igb.protannot.interproscan.api.InterProscanService$Status.valueOf(InterProscanService.java:29) ~[-1688969961404:na]
at org.lorainelab.igb.protannot.interproscan.InterProscanServiceRest.status(InterProscanServiceRest.java:106) ~[-1688969961404:na]
at org.lorainelab.igb.protannot.ProtAnnotService$5.run(ProtAnnotService.java:692) [-1688969961404:na]
at java.util.TimerThread.mainLoop(Timer.java:555) [na:1.8.0_332]
at java.util.TimerThread.run(Timer.java:505) [na:1.8.0_332]
01:21:05.130 INFO o.l.igb.protannot.ProtAnnotService - iprscan5-R20230710-072102-0693-74435734-p1m Error
For this task, determine if this error re-occurs in other searches.
For your reference, one gene model searched. The gene model was "AT1G20920.7" from the "Araport" track of Arabidopsis genome version A_thaliana_Jun_2009.
I discovered the error during final testing of IGBF-3202.
Attachments
Issue Links
Activity
| Field | Original Value | New Value |
|---|---|---|
| Epic Link | IGBF-1908 [ 17998 ] |
| Rank | Ranked higher |
| Description |
I attempted to run a search of InterPro using ProtAnnot but the search did not execute properly.
It looks like maybe the InterProScan Web service API has changed and ProtAnnot needs to be updated. The error: {quote} 01:20:10.863 INFO com.affymetrix.igb.IGB - Completed loading sequence for Chr1 : 7,284,963 - 7,290,069 from IGB Quickload http://lorainelab-quickload.scidas.org/quickload/ 01:21:04.688 INFO o.l.i.p.i.InterProscanServiceRest - iprscan5-R20230710-072102-0693-74435734-p1m 01:21:05.129 ERROR o.l.i.p.i.InterProscanServiceRest - No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED java.lang.IllegalArgumentException: No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED at java.lang.Enum.valueOf(Enum.java:238) ~[na:1.8.0_332] at org.lorainelab.igb.protannot.interproscan.api.InterProscanService$Status.valueOf(InterProscanService.java:29) ~[-1688969961404:na] at org.lorainelab.igb.protannot.interproscan.InterProscanServiceRest.status(InterProscanServiceRest.java:106) ~[-1688969961404:na] at org.lorainelab.igb.protannot.ProtAnnotService$5.run(ProtAnnotService.java:692) [-1688969961404:na] at java.util.TimerThread.mainLoop(Timer.java:555) [na:1.8.0_332] at java.util.TimerThread.run(Timer.java:505) [na:1.8.0_332] 01:21:05.130 INFO o.l.igb.protannot.ProtAnnotService - iprscan5-R20230710-072102-0693-74435734-p1m Error {quote} For this task, determine if this error re-occurs in other searches. For your reference, one gene model searched. The gene model was "AT1G20920.7" from the "Araport" track of Arabidopsis genome version A_thaliana_Jun_2009. I discovered the error during final testing of |
I attempted to run a search of InterPro using ProtAnnot but the search did not execute properly.
Note: To understand how to run a search of InterPro using ProtAnnot, watch the video linked from the top page of https://apps.bioviz.org. The first part of the video explains how to install an IGB App using the App Store. You can skip the first part, since you probably should work with the development (current master branch) version of ProtAnnot available from the Downloads section of a ProtAnnot bitbucket repository. To just see how to run a search with ProtAnnot, you can skip ahead to 1:43 minutes into the video. It looks like maybe the InterProScan Web service API has changed and ProtAnnot needs to be updated. The error: {quote} 01:20:10.863 INFO com.affymetrix.igb.IGB - Completed loading sequence for Chr1 : 7,284,963 - 7,290,069 from IGB Quickload http://lorainelab-quickload.scidas.org/quickload/ 01:21:04.688 INFO o.l.i.p.i.InterProscanServiceRest - iprscan5-R20230710-072102-0693-74435734-p1m 01:21:05.129 ERROR o.l.i.p.i.InterProscanServiceRest - No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED java.lang.IllegalArgumentException: No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED at java.lang.Enum.valueOf(Enum.java:238) ~[na:1.8.0_332] at org.lorainelab.igb.protannot.interproscan.api.InterProscanService$Status.valueOf(InterProscanService.java:29) ~[-1688969961404:na] at org.lorainelab.igb.protannot.interproscan.InterProscanServiceRest.status(InterProscanServiceRest.java:106) ~[-1688969961404:na] at org.lorainelab.igb.protannot.ProtAnnotService$5.run(ProtAnnotService.java:692) [-1688969961404:na] at java.util.TimerThread.mainLoop(Timer.java:555) [na:1.8.0_332] at java.util.TimerThread.run(Timer.java:505) [na:1.8.0_332] 01:21:05.130 INFO o.l.igb.protannot.ProtAnnotService - iprscan5-R20230710-072102-0693-74435734-p1m Error {quote} For this task, determine if this error re-occurs in other searches. For your reference, one gene model searched. The gene model was "AT1G20920.7" from the "Araport" track of Arabidopsis genome version A_thaliana_Jun_2009. I discovered the error during final testing of |
| Description |
I attempted to run a search of InterPro using ProtAnnot but the search did not execute properly.
Note: To understand how to run a search of InterPro using ProtAnnot, watch the video linked from the top page of https://apps.bioviz.org. The first part of the video explains how to install an IGB App using the App Store. You can skip the first part, since you probably should work with the development (current master branch) version of ProtAnnot available from the Downloads section of a ProtAnnot bitbucket repository. To just see how to run a search with ProtAnnot, you can skip ahead to 1:43 minutes into the video. It looks like maybe the InterProScan Web service API has changed and ProtAnnot needs to be updated. The error: {quote} 01:20:10.863 INFO com.affymetrix.igb.IGB - Completed loading sequence for Chr1 : 7,284,963 - 7,290,069 from IGB Quickload http://lorainelab-quickload.scidas.org/quickload/ 01:21:04.688 INFO o.l.i.p.i.InterProscanServiceRest - iprscan5-R20230710-072102-0693-74435734-p1m 01:21:05.129 ERROR o.l.i.p.i.InterProscanServiceRest - No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED java.lang.IllegalArgumentException: No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED at java.lang.Enum.valueOf(Enum.java:238) ~[na:1.8.0_332] at org.lorainelab.igb.protannot.interproscan.api.InterProscanService$Status.valueOf(InterProscanService.java:29) ~[-1688969961404:na] at org.lorainelab.igb.protannot.interproscan.InterProscanServiceRest.status(InterProscanServiceRest.java:106) ~[-1688969961404:na] at org.lorainelab.igb.protannot.ProtAnnotService$5.run(ProtAnnotService.java:692) [-1688969961404:na] at java.util.TimerThread.mainLoop(Timer.java:555) [na:1.8.0_332] at java.util.TimerThread.run(Timer.java:505) [na:1.8.0_332] 01:21:05.130 INFO o.l.igb.protannot.ProtAnnotService - iprscan5-R20230710-072102-0693-74435734-p1m Error {quote} For this task, determine if this error re-occurs in other searches. For your reference, one gene model searched. The gene model was "AT1G20920.7" from the "Araport" track of Arabidopsis genome version A_thaliana_Jun_2009. I discovered the error during final testing of |
I attempted to run a search of InterPro using ProtAnnot but the search did not execute properly.
It looks like maybe the InterProScan Web service API has changed and ProtAnnot needs to be updated. Note: To understand how to run a search of InterPro using ProtAnnot, watch the video linked from the top page of https://apps.bioviz.org. The first part of the video explains how to install an IGB App using the App Store. You can skip the first part, since you probably should work with the development (current master branch) version of ProtAnnot available from the Downloads section of a ProtAnnot bitbucket repository. To just see how to run a search with ProtAnnot, you can skip ahead to 1:43 minutes into the video. The error: {quote} 01:20:10.863 INFO com.affymetrix.igb.IGB - Completed loading sequence for Chr1 : 7,284,963 - 7,290,069 from IGB Quickload http://lorainelab-quickload.scidas.org/quickload/ 01:21:04.688 INFO o.l.i.p.i.InterProscanServiceRest - iprscan5-R20230710-072102-0693-74435734-p1m 01:21:05.129 ERROR o.l.i.p.i.InterProscanServiceRest - No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED java.lang.IllegalArgumentException: No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED at java.lang.Enum.valueOf(Enum.java:238) ~[na:1.8.0_332] at org.lorainelab.igb.protannot.interproscan.api.InterProscanService$Status.valueOf(InterProscanService.java:29) ~[-1688969961404:na] at org.lorainelab.igb.protannot.interproscan.InterProscanServiceRest.status(InterProscanServiceRest.java:106) ~[-1688969961404:na] at org.lorainelab.igb.protannot.ProtAnnotService$5.run(ProtAnnotService.java:692) [-1688969961404:na] at java.util.TimerThread.mainLoop(Timer.java:555) [na:1.8.0_332] at java.util.TimerThread.run(Timer.java:505) [na:1.8.0_332] 01:21:05.130 INFO o.l.igb.protannot.ProtAnnotService - iprscan5-R20230710-072102-0693-74435734-p1m Error {quote} For this task, determine if this error re-occurs in other searches. For your reference, one gene model searched. The gene model was "AT1G20920.7" from the "Araport" track of Arabidopsis genome version A_thaliana_Jun_2009. I discovered the error during final testing of |
| Description |
I attempted to run a search of InterPro using ProtAnnot but the search did not execute properly.
It looks like maybe the InterProScan Web service API has changed and ProtAnnot needs to be updated. Note: To understand how to run a search of InterPro using ProtAnnot, watch the video linked from the top page of https://apps.bioviz.org. The first part of the video explains how to install an IGB App using the App Store. You can skip the first part, since you probably should work with the development (current master branch) version of ProtAnnot available from the Downloads section of a ProtAnnot bitbucket repository. To just see how to run a search with ProtAnnot, you can skip ahead to 1:43 minutes into the video. The error: {quote} 01:20:10.863 INFO com.affymetrix.igb.IGB - Completed loading sequence for Chr1 : 7,284,963 - 7,290,069 from IGB Quickload http://lorainelab-quickload.scidas.org/quickload/ 01:21:04.688 INFO o.l.i.p.i.InterProscanServiceRest - iprscan5-R20230710-072102-0693-74435734-p1m 01:21:05.129 ERROR o.l.i.p.i.InterProscanServiceRest - No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED java.lang.IllegalArgumentException: No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED at java.lang.Enum.valueOf(Enum.java:238) ~[na:1.8.0_332] at org.lorainelab.igb.protannot.interproscan.api.InterProscanService$Status.valueOf(InterProscanService.java:29) ~[-1688969961404:na] at org.lorainelab.igb.protannot.interproscan.InterProscanServiceRest.status(InterProscanServiceRest.java:106) ~[-1688969961404:na] at org.lorainelab.igb.protannot.ProtAnnotService$5.run(ProtAnnotService.java:692) [-1688969961404:na] at java.util.TimerThread.mainLoop(Timer.java:555) [na:1.8.0_332] at java.util.TimerThread.run(Timer.java:505) [na:1.8.0_332] 01:21:05.130 INFO o.l.igb.protannot.ProtAnnotService - iprscan5-R20230710-072102-0693-74435734-p1m Error {quote} For this task, determine if this error re-occurs in other searches. For your reference, one gene model searched. The gene model was "AT1G20920.7" from the "Araport" track of Arabidopsis genome version A_thaliana_Jun_2009. I discovered the error during final testing of |
I attempted to run a search of InterPro using ProtAnnot but the search did not execute properly. (See attached image for a view of ProtAnnot.)
It looks like maybe the InterProScan Web service API has changed and ProtAnnot needs to be updated. Note: To understand how to run a search of InterPro using ProtAnnot, watch the video linked from the top page of https://apps.bioviz.org. The first part of the video explains how to install an IGB App using the App Store. You can skip the first part, since you probably should work with the development (current master branch) version of ProtAnnot available from the Downloads section of a ProtAnnot bitbucket repository. To just see how to run a search with ProtAnnot, you can skip ahead to 1:43 minutes into the video. The error: {quote} 01:20:10.863 INFO com.affymetrix.igb.IGB - Completed loading sequence for Chr1 : 7,284,963 - 7,290,069 from IGB Quickload http://lorainelab-quickload.scidas.org/quickload/ 01:21:04.688 INFO o.l.i.p.i.InterProscanServiceRest - iprscan5-R20230710-072102-0693-74435734-p1m 01:21:05.129 ERROR o.l.i.p.i.InterProscanServiceRest - No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED java.lang.IllegalArgumentException: No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED at java.lang.Enum.valueOf(Enum.java:238) ~[na:1.8.0_332] at org.lorainelab.igb.protannot.interproscan.api.InterProscanService$Status.valueOf(InterProscanService.java:29) ~[-1688969961404:na] at org.lorainelab.igb.protannot.interproscan.InterProscanServiceRest.status(InterProscanServiceRest.java:106) ~[-1688969961404:na] at org.lorainelab.igb.protannot.ProtAnnotService$5.run(ProtAnnotService.java:692) [-1688969961404:na] at java.util.TimerThread.mainLoop(Timer.java:555) [na:1.8.0_332] at java.util.TimerThread.run(Timer.java:505) [na:1.8.0_332] 01:21:05.130 INFO o.l.igb.protannot.ProtAnnotService - iprscan5-R20230710-072102-0693-74435734-p1m Error {quote} For this task, determine if this error re-occurs in other searches. For your reference, one gene model searched. The gene model was "AT1G20920.7" from the "Araport" track of Arabidopsis genome version A_thaliana_Jun_2009. I discovered the error during final testing of |
| Description |
I attempted to run a search of InterPro using ProtAnnot but the search did not execute properly. (See attached image for a view of ProtAnnot.)
It looks like maybe the InterProScan Web service API has changed and ProtAnnot needs to be updated. Note: To understand how to run a search of InterPro using ProtAnnot, watch the video linked from the top page of https://apps.bioviz.org. The first part of the video explains how to install an IGB App using the App Store. You can skip the first part, since you probably should work with the development (current master branch) version of ProtAnnot available from the Downloads section of a ProtAnnot bitbucket repository. To just see how to run a search with ProtAnnot, you can skip ahead to 1:43 minutes into the video. The error: {quote} 01:20:10.863 INFO com.affymetrix.igb.IGB - Completed loading sequence for Chr1 : 7,284,963 - 7,290,069 from IGB Quickload http://lorainelab-quickload.scidas.org/quickload/ 01:21:04.688 INFO o.l.i.p.i.InterProscanServiceRest - iprscan5-R20230710-072102-0693-74435734-p1m 01:21:05.129 ERROR o.l.i.p.i.InterProscanServiceRest - No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED java.lang.IllegalArgumentException: No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED at java.lang.Enum.valueOf(Enum.java:238) ~[na:1.8.0_332] at org.lorainelab.igb.protannot.interproscan.api.InterProscanService$Status.valueOf(InterProscanService.java:29) ~[-1688969961404:na] at org.lorainelab.igb.protannot.interproscan.InterProscanServiceRest.status(InterProscanServiceRest.java:106) ~[-1688969961404:na] at org.lorainelab.igb.protannot.ProtAnnotService$5.run(ProtAnnotService.java:692) [-1688969961404:na] at java.util.TimerThread.mainLoop(Timer.java:555) [na:1.8.0_332] at java.util.TimerThread.run(Timer.java:505) [na:1.8.0_332] 01:21:05.130 INFO o.l.igb.protannot.ProtAnnotService - iprscan5-R20230710-072102-0693-74435734-p1m Error {quote} For this task, determine if this error re-occurs in other searches. For your reference, one gene model searched. The gene model was "AT1G20920.7" from the "Araport" track of Arabidopsis genome version A_thaliana_Jun_2009. I discovered the error during final testing of |
I attempted to run a search of InterPro using ProtAnnot but the search did not execute properly. (See attached image for a view of ProtAnnot.)
It looks like maybe the InterProScan Web service API has changed and ProtAnnot needs to be updated. Note: To understand how to run a search of InterPro using ProtAnnot, watch the video linked from the top page of https://apps.bioviz.org. The first part of the video explains how to install an IGB App using the App Store. You can skip the first part, since you probably should work with the development (current master branch) version of ProtAnnot available from the Downloads section of a ProtAnnot bitbucket repository. To just see how to run a search with ProtAnnot, you can skip ahead to 1:43 minutes into the video. The error, copied from IGB "log" tab: {quote} 01:20:10.863 INFO com.affymetrix.igb.IGB - Completed loading sequence for Chr1 : 7,284,963 - 7,290,069 from IGB Quickload http://lorainelab-quickload.scidas.org/quickload/ 01:21:04.688 INFO o.l.i.p.i.InterProscanServiceRest - iprscan5-R20230710-072102-0693-74435734-p1m 01:21:05.129 ERROR o.l.i.p.i.InterProscanServiceRest - No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED java.lang.IllegalArgumentException: No enum constant org.lorainelab.igb.protannot.interproscan.api.InterProscanService.Status.QUEUED at java.lang.Enum.valueOf(Enum.java:238) ~[na:1.8.0_332] at org.lorainelab.igb.protannot.interproscan.api.InterProscanService$Status.valueOf(InterProscanService.java:29) ~[-1688969961404:na] at org.lorainelab.igb.protannot.interproscan.InterProscanServiceRest.status(InterProscanServiceRest.java:106) ~[-1688969961404:na] at org.lorainelab.igb.protannot.ProtAnnotService$5.run(ProtAnnotService.java:692) [-1688969961404:na] at java.util.TimerThread.mainLoop(Timer.java:555) [na:1.8.0_332] at java.util.TimerThread.run(Timer.java:505) [na:1.8.0_332] 01:21:05.130 INFO o.l.igb.protannot.ProtAnnotService - iprscan5-R20230710-072102-0693-74435734-p1m Error {quote} For this task, determine if this error re-occurs in other searches. For your reference, one gene model searched. The gene model was "AT1G20920.7" from the "Araport" track of Arabidopsis genome version A_thaliana_Jun_2009. I discovered the error during final testing of |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Assignee | Kaushik Gopu [ kgopu ] |
| Attachment | Dscam1.png [ 17915 ] |
| Status | In Progress [ 3 ] | Needs 1st Level Review [ 10005 ] |
| Assignee | Kaushik Gopu [ kgopu ] | Ann Loraine [ aloraine ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Status | First Level Review in Progress [ 10301 ] | To-Do [ 10305 ] |
| Assignee | Ann Loraine [ aloraine ] | Kaushik Gopu [ kgopu ] |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Status | In Progress [ 3 ] | Needs 1st Level Review [ 10005 ] |
| Assignee | Kaushik Gopu [ kgopu ] | Nowlan Freese [ nfreese ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Status | First Level Review in Progress [ 10301 ] | Ready for Pull Request [ 10304 ] |
| Assignee | Nowlan Freese [ nfreese ] | Kaushik Gopu [ kgopu ] |
| Status | Ready for Pull Request [ 10304 ] | Pull Request Submitted [ 10101 ] |
| Assignee | Kaushik Gopu [ kgopu ] | Ann Loraine [ aloraine ] |
| Status | Pull Request Submitted [ 10101 ] | Reviewing Pull Request [ 10303 ] |
| Status | Reviewing Pull Request [ 10303 ] | Merged Needs Testing [ 10002 ] |
| Assignee | Ann Loraine [ aloraine ] |
| Status | Merged Needs Testing [ 10002 ] | Post-merge Testing In Progress [ 10003 ] |
| Assignee | Nowlan Freese [ nfreese ] |
| Assignee | Nowlan Freese [ nfreese ] | Kaushik Gopu [ kgopu ] |
| Resolution | Done [ 10000 ] | |
| Status | Post-merge Testing In Progress [ 10003 ] | Closed [ 6 ] |
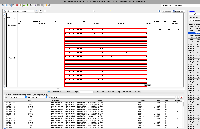
Tested on IGB 9.1.10 release on Mac
Ran ProtAnnot and saw the error mentioned in the description as well as the following error.
Note: I ran ProtAnnot on two gene models, one Finished and one had a status of Error.
Exception in thread "AWT-EventQueue-0" java.lang.NullPointerException at org.lorainelab.igb.protannot.model.InterProScanTableModel$InterProScanTableData.access$100(InterProScanTableModel.java:113) at org.lorainelab.igb.protannot.model.InterProScanTableModel.getValueAt(InterProScanTableModel.java:90) at javax.swing.JTable.getValueAt(JTable.java:2720) at com.jidesoft.grid.JideTable.getValueAt(Unknown Source) at com.jidesoft.grid.JideTable.prepareRenderer(Unknown Source) at com.affymetrix.igb.swing.jide.StyledJTable.prepareRenderer(StyledJTable.java:137) at com.jidesoft.plaf.basic.BasicJideTableUIDelegate.paintCell(Unknown Source) at com.jidesoft.plaf.basic.BasicJideTableUIDelegate.a(Unknown Source) at com.jidesoft.plaf.basic.BasicJideTableUIDelegate.paint(Unknown Source) at com.jidesoft.plaf.aqua.AquaJideTableUI.paint(Unknown Source) at javax.swing.plaf.ComponentUI.update(ComponentUI.java:161) at javax.swing.JComponent.paintComponent(JComponent.java:780) at javax.swing.JComponent.paint(JComponent.java:1056) at javax.swing.JComponent.paintToOffscreen(JComponent.java:5210) at javax.swing.RepaintManager$PaintManager.paintDoubleBuffered(RepaintManager.java:1579) at javax.swing.RepaintManager$PaintManager.paint(RepaintManager.java:1502) at javax.swing.RepaintManager.paint(RepaintManager.java:1272) at javax.swing.JComponent._paintImmediately(JComponent.java:5158) at javax.swing.JComponent.paintImmediately(JComponent.java:4969) at javax.swing.RepaintManager$4.run(RepaintManager.java:831) at javax.swing.RepaintManager$4.run(RepaintManager.java:814) at java.security.AccessController.doPrivileged(Native Method) at java.security.ProtectionDomain$JavaSecurityAccessImpl.doIntersectionPrivilege(ProtectionDomain.java:74) at javax.swing.RepaintManager.paintDirtyRegions(RepaintManager.java:814) at javax.swing.RepaintManager.paintDirtyRegions(RepaintManager.java:789) at javax.swing.RepaintManager.prePaintDirtyRegions(RepaintManager.java:738) at javax.swing.RepaintManager.access$1200(RepaintManager.java:64) at javax.swing.RepaintManager$ProcessingRunnable.run(RepaintManager.java:1732) at java.awt.event.InvocationEvent.dispatch(InvocationEvent.java:311) at java.awt.EventQueue.dispatchEventImpl(EventQueue.java:758) at java.awt.EventQueue.access$500(EventQueue.java:97) at java.awt.EventQueue$3.run(EventQueue.java:709) at java.awt.EventQueue$3.run(EventQueue.java:703) at java.security.AccessController.doPrivileged(Native Method) at java.security.ProtectionDomain$JavaSecurityAccessImpl.doIntersectionPrivilege(ProtectionDomain.java:74) at java.awt.EventQueue.dispatchEvent(EventQueue.java:728) at java.awt.EventDispatchThread.pumpOneEventForFilters(EventDispatchThread.java:205) at java.awt.EventDispatchThread.pumpEventsForFilter(EventDispatchThread.java:116) at java.awt.EventDispatchThread.pumpEventsForHierarchy(EventDispatchThread.java:105) at java.awt.EventDispatchThread.pumpEvents(EventDispatchThread.java:101) at java.awt.EventDispatchThread.pumpEvents(EventDispatchThread.java:93) at java.awt.EventDispatchThread.run(EventDispatchThread.java:82)