Details
-
Type:
Task
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: 10.1.0
-
Labels:None
-
Story Points:2
-
Epic Link:
-
Sprint:Spring 6, Spring 7
Description
Develop code to connect to this https://api.genome.ucsc.edu/getData/track?genome=hg38&track=bloodHaoCellType&chrom=chr1&start=1&end=2480000 API and load Data for the barChart file type.
Investigate whether this file format can be parsed as a bed or graph and add logic to parse the barChart file format.
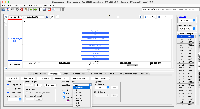
The list of tracks for the hg38 genome has the sample names for bigBarChart listed as barChartBars: https://api.genome.ucsc.edu/list/tracks?genome=hg38;trackLeavesOnly=1
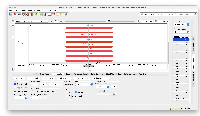
It also has the colors which is interesting, as barChartColors. If we wanted to really go in deep on this one, we could make a bed file that is color coded using their provided colors. This might be a lot of work, so might not be worth it, but it would be neat if we made each cell type its own line in the bed file. I attached an example file, bloodHaoCellType.bed, to show what this could look like in IGB. Compare to the UCSC.