Details
-
Type:
Task
-
Status: Merged Needs Testing (View Workflow)
-
Priority:
Blocker
-
Resolution: Unresolved
-
Affects Version/s: None
-
Fix Version/s: None
-
Labels:None
-
Story Points:5
-
Epic Link:
-
Sprint:Spring 6, Spring 7, Spring 8, Spring 9, Spring 10
Description
Pass the schema of the UCSC data in the properties field while constructing the dataset. This enables developers to extract the schema/metadata of the data easily. This metadata can be used to provide rich set of Filter by/Color by options to IGB users. Structure the metadata in the same way as actual data, for example, [attribute 1, attribute 2, ..., props].
Attachments
- Screenshot_1.png
- 211 kB
- Screenshot_2.png
- 64 kB
- Screenshot_3.png
- 118 kB
- Screenshot_4.png
- 72 kB
Issue Links
- is blocked by
-
IGBF-3640 Add expand color by and Filter by to include the props
-
- Reviewing Pull Request
-
Activity
After going through the code, found out that the featureProps is actually a Map<String, String> variable and won't be able to store a map object as a value. Checking with Kaushik Gopu whether we can change the variable to Map<String, Object> type or have the props stored in any of the below ways:
1.
{
"genome": "galGal6",
"track": "nestedRepeats",
"id": "number",
"repClass": "string",
"repFamily": "string"
}
2.
{
"genome": "galGal6",
"track": "nestedRepeats",
"props": "id,repClass,repFamily"
}
3.
{
"genome": "galGal6",
"track": "nestedRepeats",
"props": "id:number,repClass:string,repFamily:string"
}
Jaya Sravani Sirigineedi, the third option looks good to me as it contains information about data and its type. Please proceed with the implementation. About changing String to Object, Let's not modify any of the existing code, as we don't know the impact of the change.
Done changes to code to send the schema info like the 3rd option mentioned above. Modified the test case accordingly. Code changes are available at the branch: https://bitbucket.org/jaya-sravani/integrated-genome-browser/branch/IGBF-3660.
To Test:
As this is just a variable addition, you won't be able to see it in IGB, instead can be found in the variables while debugging. Follow the below steps to debug:
- Build the jar file for the branch .
- Keep a debug point at line 136 in the RestApiDataProvider class.
- Start the application in debug mode and select any species.
- Now while loading the tracks, your IDE should change to debug view and show some value like the below in the featureProps variable for bed tracks, it should filter out the bed properties and append only the additional columns.
{
"genome": "allMis1",
"track": "cpgIslandExt",
"props": "bin:number,length:number,cpgNum:number,gcNum:number,perCpg:double,perGc:double,obsExp:double"
}
Kaushik Gopu please check and let me know if there's any change required.
- There is an intermittent issue that returns null when the code is trying to get props from the featureProps. This issue persists throughout the session, meaning that the user does not have access to props for the Color-By operation. Initially, I thought the problem was with the external service we were using to get the metadata. To test this, I have added a log statement that logs the variable featureProps after a call to the REST service (in between line 137 and line 138 of the RestApiDataProvider class). Every single time I ran the application, I could see the props appear in the logs. However, for some reason, these props are passed as null (check line 137 of RestApiDataProvider class) though the data exists.
- Screenshot_1 and Screenshot_2: Props appeared in the logs for the dataset cytoBandIdeo, but they are pointed to null in Screenshot_2.
- Screenshot_3 and Screenshot_4: Props appeared in the logs for the dataset cpgIslandExt, and the props are available in Screenshot_4.
- Screenshot_2 and Screenshot_4 are taken while debugging the application. This code is for to get the props data for the selected dataset.
- This issue is not associated with any particular dataset. Sometimes, cpgIslandExt does not have props data, while cytoBandIdeo does, and I do not see this issue at all seven out of ten times.
After investigating the issue with Kaushik Gopu, we found the issue is not in adding the props code but while retrieving it, in the code there is a condition to filter the selected datasets from all the available datasets and from the result it is considering the first one which is giving a problem as right now we have both UCSC Das datasets and UCSC Rest datasets available, so for a particular track you might get two results from it and sometimes it might return the Rest one or the Das one. Because of this, the getProperties() is returning null, as we have the additional properties added only to the Rest tracks. This is can be resolved by checking the DataProvider as well in the condition.
Raised a PR https://bitbucket.org/lorainelab/integrated-genome-browser/pull-requests/995 to merge the changes for the schema addition. Kaushik Gopu please review and let me know if there's any issue.
PR is merged.
I'm having a lot of trouble loading the human genome with this commit. If I go back to a previous commit without IGBF-3660 I am able to load the human genome quickly. I'm wondering if we are making a lot of API calls all at once. I also think we may need some logging.
Jaya Sravani Sirigineedi - let's discuss options once you have time.
Moving back to to-do
Yes, Nowlan Freese. I observed it just before sometime while testing the barChart type. I wanted to speak with you too. I am already working on it. When implementing this code to load schema, we didn't add the GenePred, narrowPeak, and barChart ones. After adding these, the number of tracks has been increased and the schema API is being called for all the tracks, I am trying to optimize it right now, and will give a revised PR by EOD.
There was a JsonParsing error the itemCount jsonElement was mapped to int before and when the other file types were added, for a few of them it had a very large number as a value which resulted in the parsing error, and because of that IGB got stuck and wasn't able to load the data. This is fixed and also, we observed that there are a lot of tracks so added logic to call the schema API only when the track is in any one of these formats: ("bed", "bigbed", "beddetail", "biggenepred"), after doing this change the statistics are as below:
No of times schema API is called: 357 and time taken for execution: 129671 (nearly 2 minutes)
When i commented the code to call the schema API then the time for execution is:
No of times schema API is called: 0 and time taken for execution: 1165, about second
which still is a lot of calls made to the UCSC server at a time when the user launches the app. If we can call the schema API when the user clicks on a particular track that would become a lot more efficient. Kaushik Gopu and Nowlan Freese let's discuss about this when you have time. Below is the updated code with the above mentioned changes: https://bitbucket.org/jaya-sravani/integrated-genome-browser/branch/IGBF-3659
Raised a PR for the updated code: https://bitbucket.org/lorainelab/integrated-genome-browser/pull-requests/997. Nowlan Freese Please review the PR and let me know if there are any issues.
PR is now merged. Thank you Jaya Sravani Sirigineedi for letting me know you were waiting on this!
After discussion with Sravani, we have decided to attempt to only make the schema API call when a user selects a specific data set.
For example, a user selects the galGal6 genome (no schema API calls are made). Then, under Available Data > UCSC REST (UCSC REST), when the user selects cpgIslandExt the schema API call is made for just cpgIslandExt.
Started investigating on where to add the integration of this schema API i.e., have to find what part of code is responsible for displaying the empty track when user clicks on the track checkbox under Available Data.
When the user clicks on the track checkbox under Available Data, the addFeatureTier method in GeneralLoadUtils is being called which uses the addEmptyTierFor method from TrackView class to display both the forward and reverse empty tracks. Code can be added here to get the schema for the selected track as the Dataset is available in the method. But the only problem is right now in Kaushik Gopu implementation he is getting tracks from the DataProvider again, this might override the featureProps and assign it to null, have to chack whether the same dataset can be used in his implementation as this is saved in a ITrackStyleExtended instance as well.
Had a discussion with Kaushik Gopu and we think the above approach might work, implementing it right now. Will update here once it's tested.
Completed integrating the schema API in the addEmptyTierFor() method code which is used to load the empty tier when the user clicks on a track, added the ucsc-rest-api-service module to achieve this but faced a few runtime errors when adding the dependency and had to add the required packages in the export package section in the pom.xml to fix the errors. Also, the addEmptyTierFor method is being called a few other times too, so added a condition so that the API call is made only once and not every time when this method is called.
Working on using this populated props in the ColorBy and FilterBy options. https://jira.bioviz.org/browse/IGBF-3640 once this story is also tested then will push the code as both go hand-in-hand.
Development is completed, tested the code and it is working as expected. Updated code is at the branch: https://bitbucket.org/jaya-sravani/integrated-genome-browser/branch/IGBF-3660.
To Test:
- Build the jar file for the branch.
- Keep a debug point at line 238 in the TrackView class.
- Start the application in debug mode, select any species, and select any one of the tracks from the available ones.
- Now while loading the empty track, your IDE should change to debug view and show some value like the below in the featureProps variable for bed tracks, it should filter out the bed properties and append only the additional columns.
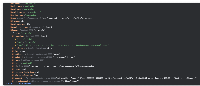
Need to do a small change as the code for this ticket https://jira.bioviz.org/browse/IGBF-3703 adds a description property to the featureprops and it impacts the modified code for this ticket. Will update it and push the code.
Updated the code and it's working as expected. Code is available at:https://bitbucket.org/jaya-sravani/integrated-genome-browser/branch/IGBF-3660
Built Sravani's branch locally on my Mac.
Schema is now fetched only when the user selects a specific data set from the UCSC REST in the Available Data panel. Schema is passed to props and appears correctly when using Color By or Filter By.
Ready for pull request.
Created a single Pull request for both tickets, https://jira.bioviz.org/browse/IGBF-3660 and https://jira.bioviz.org/browse/IGBF-3640. Nowlan Freese already reviewed it, Kaushik Gopu Please review it and let me know if find any issues.
Suggested surrounding code with try-catch block to avoid unexpected failures while retrieving props data. Other than that, everything looks great to me.
PR is merged and new installers are built and deployed to bioviz.org "early access" section.
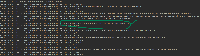
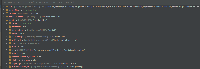

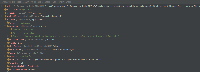
As per discussion with Kaushik Gopu, will add the asked columns in the featureProps variable in DataSet objects created while getting the available datasets using the schema response from the schema API, example API: https://api.genome.ucsc.edu/list/schema?genome=galGal6;track=nestedRepeats. Below is an example showing how the featureProps look like:
Example schema response:
featureProps for the above schema:
{ "genome": "galGal6", "track": "nestedRepeats", "props": { "id": "number", "repClass": "string", "repFamily": "string" } }