Details
-
Type:
Task
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: 10.1.0
-
Labels:None
-
Story Points:2
-
Epic Link:
-
Sprint:Spring 7, Spring 8, Spring 9
Description
It is very tough for a user to search for a track in the available tracks panel as there are a lot of tracks in the hg38 genome so we have to investigate a way to make folders for each file type inside the UCSC Rest folder so it becomes a little bit easier for the user to search a specific track.
Attachments
Issue Links
- relates to
-
IGBF-3926 Organize UCSC data by "group" and "subgroup"
-
- Closed
-
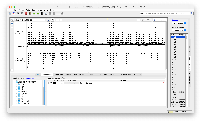
Investigated and found the code on how to make folders, changed the code in UCSC Rest as well to make the folders. Nowlan Freese Need your input on on how to organise the folders. Is it okay if I do folders for all the file types or do you want me to make folders for only the few file types that have a lot of tracks. Please review the below screenshot.
