I was able to add back useq support in IGB by reverting the commit from IGBF-3146 and then removing some obsolete code (see this post) and updating some import statements.
Branch: https://bitbucket.org/nfreese/nowlanfork-igb/branch/IGBF-3705
I used useq test files (attached) that are included in the IGB code base from core/genometry/src/test/resources/data/useq
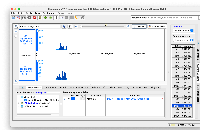
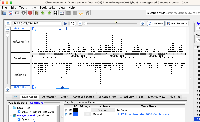
While testing I noticed that loading the test file chr17_H_sapiens_Mar_2006_Graph.useq caused the track name to appear multiple times in the Data Management Table (see useqBroken.png). When attempting to delete the "extra" tracks, I see the following error:
java.lang.NullPointerException: Cannot invoke "com.affymetrix.genometry.general.DataSet.getDataSetName()" because the return value of "com.affymetrix.genometry.style.ITrackStyleExtended.getFeature()" is null
This same behavior does not appear in the last version of IGB that supports useq (9.1.8). I was able to track down the cause of this behavior to a change made in IGBF-3584 (commit). The change to DataSet.java on line 253 appears to cause the issue, as reverting that specific edit appears to "fix" the issue for the useq file (see useqWorking.png).
However, the same issue does not appear for the file chr17_H_sapiens_Mar_2006_Region.useq, which loads as an annotation (in comparison to chr17_H_sapiens_Mar_2006_Graph.useq, which loads as a graph). As I am unfamiliar with the useq file format it is unclear why this issue is occurring. As we have seen no other issues with loading graph files, the issue may be a quirk of useq graph files.
To reproduce the issue:
- Fetch my branch
- Open the H_sapiens_Mar_2006 genome (it is an older human genome version)
- Navigate to chr17
- Add and load the test files (attached)
- Look for additional tracks to appear
- Attempt to delete the additional tracks
Task
Major
IGBF-3146 Remove "useq" packages from genometry bundle in IGB
User conversation: