Details
-
Type:
Task
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: None
-
Labels:None
-
Story Points:3
-
Sprint:Summer 2, Summer 6, Summer 7, Fall 3, Fall 4, Fall 6, Fall 7, Winter 1
Description
Situation: We have identified a publicly available Nebula Genomics CRAM file (link). Nowlan has downloaded the file and created an index, and was able to view the file in IGB. We would like to make this file available to IGB users as an example of a consumer genomics CRAM file.
Task: Upload and deploy the CRAM file (NB72462M.cram), CRAM index, VCF file, and VCF index and make them available through an IGB Quickload. Also deploy the IGBF-3841 via the annots.xml.
Note: Data are from Personal Genome Project
Link to metadata in PGP: https://my.pgp-hms.org/profile/huF7A4DE
Link to PGP search page: https://my.pgp-hms.org/public_genetic_data
Revision:
- Scope increase - include Nebula Genomics and Sequence.com data files for Donor 1
- Scope increase - explore data in region of BRCA1 as preliminary use case, beginning tutorial
Attachments
Issue Links
Activity
| Field | Original Value | New Value |
|---|---|---|
| Epic Link | IGBF-1765 [ 17855 ] |
| Link | This issue relates to IGBF-3642 [ IGBF-3642 ] |
| Sprint | Summer 1 [ 195 ] |
| Sprint | Summer 2 [ 196 ] |
| Rank | Ranked higher |
| Sprint | Summer 2 [ 196 ] | Summer 2, Summer 3 [ 196, 197 ] |
| Rank | Ranked higher |
| Sprint | Summer 2, Summer 3 [ 196, 197 ] | Summer 2 [ 196 ] |
| Sprint | Summer 2 [ 196 ] | Summer 2, Summer 4 [ 196, 198 ] |
| Rank | Ranked higher |
| Sprint | Summer 2, Summer 4 [ 196, 198 ] | Summer 2 [ 196 ] |
| Rank | Ranked lower |
| Sprint | Summer 2 [ 196 ] | Summer 2, Summer 6 [ 196, 200 ] |
| Assignee | Paige Kulzer [ pkulzer ] |
| Assignee | Paige Kulzer [ pkulzer ] | Nowlan Freese [ nfreese ] |
| Attachment | quickload.zip [ 18462 ] |
| Description |
Situation: We have identified a publicly available Nebula Genomics CRAM file ([link|https://e4d05c260dbc25ea5821e03eb009a2b7-26238.collections.ac2it.arvadosapi.com/_/]). Nowlan has downloaded the file and created an index, and was able to view the file in IGB. We would like to make this file available to IGB users as an example of a consumer genomics CRAM file.
Task: Upload and deploy the CRAM file (NB72462M.cram), CRAM index, VCF file, and VCF index and make them available through an IGB Quickload. |
Situation: We have identified a publicly available Nebula Genomics CRAM file ([link|https://e4d05c260dbc25ea5821e03eb009a2b7-26238.collections.ac2it.arvadosapi.com/_/]). Nowlan has downloaded the file and created an index, and was able to view the file in IGB. We would like to make this file available to IGB users as an example of a consumer genomics CRAM file.
Task: Upload and deploy the CRAM file (NB72462M.cram), CRAM index, VCF file, and VCF index and make them available through an IGB Quickload. Note: Data are from Personal Genome Project Link to metadata in PGP: https://my.pgp-hms.org/profile/huF7A4DE Link to PGP search page: https://my.pgp-hms.org/public_genetic_data?utf8=%E2%9C%93&data_type=&commit=Search |
| Assignee | Nowlan Freese [ nfreese ] |
| Summary | Deploy Nebula Genomics CRAM file to web | Add individual genome CRAM file to IGB quickload via svn |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Assignee | Nowlan Freese [ nfreese ] |
| Sprint | Summer 2, Summer 6 [ 196, 200 ] | Summer 2, Summer 6, Summer 7 [ 196, 200, 201 ] |
| Rank | Ranked higher |
| Link | This issue blocks IGBF-3854 [ IGBF-3854 ] |
| Status | In Progress [ 3 ] | To-Do [ 10305 ] |
| Description |
Situation: We have identified a publicly available Nebula Genomics CRAM file ([link|https://e4d05c260dbc25ea5821e03eb009a2b7-26238.collections.ac2it.arvadosapi.com/_/]). Nowlan has downloaded the file and created an index, and was able to view the file in IGB. We would like to make this file available to IGB users as an example of a consumer genomics CRAM file.
Task: Upload and deploy the CRAM file (NB72462M.cram), CRAM index, VCF file, and VCF index and make them available through an IGB Quickload. Note: Data are from Personal Genome Project Link to metadata in PGP: https://my.pgp-hms.org/profile/huF7A4DE Link to PGP search page: https://my.pgp-hms.org/public_genetic_data?utf8=%E2%9C%93&data_type=&commit=Search |
Situation: We have identified a publicly available Nebula Genomics CRAM file ([link|https://e4d05c260dbc25ea5821e03eb009a2b7-26238.collections.ac2it.arvadosapi.com/_/]). Nowlan has downloaded the file and created an index, and was able to view the file in IGB. We would like to make this file available to IGB users as an example of a consumer genomics CRAM file.
Task: Upload and deploy the CRAM file (NB72462M.cram), CRAM index, VCF file, and VCF index and make them available through an IGB Quickload. Note: Data are from Personal Genome Project Link to metadata in PGP: https://my.pgp-hms.org/profile/huF7A4DE Link to PGP search page: https://my.pgp-hms.org/public_genetic_data |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Status | In Progress [ 3 ] | To-Do [ 10305 ] |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Attachment | quickload_V2.zip [ 18473 ] |
| Assignee | Nowlan Freese [ nfreese ] | Ann Loraine [ aloraine ] |
| Status | In Progress [ 3 ] | To-Do [ 10305 ] |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Status | In Progress [ 3 ] | Needs 1st Level Review [ 10005 ] |
| Assignee | Ann Loraine [ aloraine ] | Nowlan Freese [ nfreese ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Status | First Level Review in Progress [ 10301 ] | Needs 1st Level Review [ 10005 ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Status | First Level Review in Progress [ 10301 ] | To-Do [ 10305 ] |
| Description |
Situation: We have identified a publicly available Nebula Genomics CRAM file ([link|https://e4d05c260dbc25ea5821e03eb009a2b7-26238.collections.ac2it.arvadosapi.com/_/]). Nowlan has downloaded the file and created an index, and was able to view the file in IGB. We would like to make this file available to IGB users as an example of a consumer genomics CRAM file.
Task: Upload and deploy the CRAM file (NB72462M.cram), CRAM index, VCF file, and VCF index and make them available through an IGB Quickload. Note: Data are from Personal Genome Project Link to metadata in PGP: https://my.pgp-hms.org/profile/huF7A4DE Link to PGP search page: https://my.pgp-hms.org/public_genetic_data |
Situation: We have identified a publicly available Nebula Genomics CRAM file ([link|https://e4d05c260dbc25ea5821e03eb009a2b7-26238.collections.ac2it.arvadosapi.com/_/]). Nowlan has downloaded the file and created an index, and was able to view the file in IGB. We would like to make this file available to IGB users as an example of a consumer genomics CRAM file.
Task: Upload and deploy the CRAM file (NB72462M.cram), CRAM index, VCF file, and VCF index and make them available through an IGB Quickload. Also deploy the Note: Data are from Personal Genome Project Link to metadata in PGP: https://my.pgp-hms.org/profile/huF7A4DE Link to PGP search page: https://my.pgp-hms.org/public_genetic_data |
| Sprint | Summer 2, Summer 6, Summer 7 [ 196, 200, 201 ] | Summer 2, Summer 6, Summer 7, Fall 1 [ 196, 200, 201, 202 ] |
| Rank | Ranked higher |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Attachment | annots.xml [ 18485 ] |
| Status | In Progress [ 3 ] | To-Do [ 10305 ] |
| Attachment | quickload_v3.zip [ 18486 ] |
| Sprint | Summer 2, Summer 6, Summer 7, Fall 1 [ 196, 200, 201, 202 ] | Summer 2, Summer 6, Summer 7, Fall 2 [ 196, 200, 201, 203 ] |
| Sprint | Summer 2, Summer 6, Summer 7, Fall 2 [ 196, 200, 201, 203 ] | Summer 2, Summer 6, Summer 7, Fall 3 [ 196, 200, 201, 204 ] |
| Sprint | Summer 2, Summer 6, Summer 7, Fall 3 [ 196, 200, 201, 204 ] | Summer 2, Summer 6, Summer 7, Fall 3, Fall 4 [ 196, 200, 201, 204, 205 ] |
| Rank | Ranked higher |
| Assignee | Nowlan Freese [ nfreese ] | Ann Loraine [ aloraine ] |
| Sprint | Summer 2, Summer 6, Summer 7, Fall 3, Fall 4 [ 196, 200, 201, 204, 205 ] | Summer 2, Summer 6, Summer 7, Fall 3, Fall 4, Fall 5 [ 196, 200, 201, 204, 205, 206 ] |
| Rank | Ranked higher |
| Sprint | Summer 2, Summer 6, Summer 7, Fall 3, Fall 4, Fall 5 [ 196, 200, 201, 204, 205, 206 ] | Summer 2, Summer 6, Summer 7, Fall 3, Fall 4, Fall 6 [ 196, 200, 201, 204, 205, 207 ] |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Sprint | Summer 2, Summer 6, Summer 7, Fall 3, Fall 4, Fall 6 [ 196, 200, 201, 204, 205, 207 ] | Summer 2, Summer 6, Summer 7, Fall 3, Fall 4, Fall 6, Fall 7 [ 196, 200, 201, 204, 205, 207, 208 ] |
| Rank | Ranked higher |
| Sprint | Summer 2, Summer 6, Summer 7, Fall 3, Fall 4, Fall 6, Fall 7 [ 196, 200, 201, 204, 205, 207, 208 ] | Summer 2, Summer 6, Summer 7, Fall 3, Fall 4, Fall 6, Fall 7, Fall 8 [ 196, 200, 201, 204, 205, 207, 208, 209 ] |
| Rank | Ranked higher |
| Description |
Situation: We have identified a publicly available Nebula Genomics CRAM file ([link|https://e4d05c260dbc25ea5821e03eb009a2b7-26238.collections.ac2it.arvadosapi.com/_/]). Nowlan has downloaded the file and created an index, and was able to view the file in IGB. We would like to make this file available to IGB users as an example of a consumer genomics CRAM file.
Task: Upload and deploy the CRAM file (NB72462M.cram), CRAM index, VCF file, and VCF index and make them available through an IGB Quickload. Also deploy the Note: Data are from Personal Genome Project Link to metadata in PGP: https://my.pgp-hms.org/profile/huF7A4DE Link to PGP search page: https://my.pgp-hms.org/public_genetic_data |
Situation: We have identified a publicly available Nebula Genomics CRAM file ([link|https://e4d05c260dbc25ea5821e03eb009a2b7-26238.collections.ac2it.arvadosapi.com/_/]). Nowlan has downloaded the file and created an index, and was able to view the file in IGB. We would like to make this file available to IGB users as an example of a consumer genomics CRAM file.
Task: Upload and deploy the CRAM file (NB72462M.cram), CRAM index, VCF file, and VCF index and make them available through an IGB Quickload. Also deploy the Note: Data are from Personal Genome Project Link to metadata in PGP: https://my.pgp-hms.org/profile/huF7A4DE Link to PGP search page: https://my.pgp-hms.org/public_genetic_data Revision: * Scope increase - include Nebula Genomics and Sequence.com data files for Donor 1 |
| Description |
Situation: We have identified a publicly available Nebula Genomics CRAM file ([link|https://e4d05c260dbc25ea5821e03eb009a2b7-26238.collections.ac2it.arvadosapi.com/_/]). Nowlan has downloaded the file and created an index, and was able to view the file in IGB. We would like to make this file available to IGB users as an example of a consumer genomics CRAM file.
Task: Upload and deploy the CRAM file (NB72462M.cram), CRAM index, VCF file, and VCF index and make them available through an IGB Quickload. Also deploy the Note: Data are from Personal Genome Project Link to metadata in PGP: https://my.pgp-hms.org/profile/huF7A4DE Link to PGP search page: https://my.pgp-hms.org/public_genetic_data Revision: * Scope increase - include Nebula Genomics and Sequence.com data files for Donor 1 |
Situation: We have identified a publicly available Nebula Genomics CRAM file ([link|https://e4d05c260dbc25ea5821e03eb009a2b7-26238.collections.ac2it.arvadosapi.com/_/]). Nowlan has downloaded the file and created an index, and was able to view the file in IGB. We would like to make this file available to IGB users as an example of a consumer genomics CRAM file.
Task: Upload and deploy the CRAM file (NB72462M.cram), CRAM index, VCF file, and VCF index and make them available through an IGB Quickload. Also deploy the Note: Data are from Personal Genome Project Link to metadata in PGP: https://my.pgp-hms.org/profile/huF7A4DE Link to PGP search page: https://my.pgp-hms.org/public_genetic_data Revision: * Scope increase - include Nebula Genomics and Sequence.com data files for Donor 1 * Scope increase - explore data in region of BRCA1 as preliminary use case, beginning tutorial |
| Attachment | quickload.zip [ 18462 ] |
| Attachment | quickload_V2.zip [ 18473 ] |
| Attachment | rs80357336.png [ 18598 ] |
| Attachment | chrEBV_error.txt [ 18599 ] |
| Attachment | BothFiles-Donor1-BRCA1.png [ 18600 ] |
| Attachment | BRCA1-chr17-43092417.png [ 18606 ] |
| Status | In Progress [ 3 ] | Needs 1st Level Review [ 10005 ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Status | First Level Review in Progress [ 10301 ] | Ready for Pull Request [ 10304 ] |
| Status | Ready for Pull Request [ 10304 ] | Pull Request Submitted [ 10101 ] |
| Status | Pull Request Submitted [ 10101 ] | Reviewing Pull Request [ 10303 ] |
| Status | Reviewing Pull Request [ 10303 ] | Merged Needs Testing [ 10002 ] |
| Assignee | Ann Loraine [ aloraine ] |
| Assignee | Paige Kulzer [ pkulzer ] |
| Status | Merged Needs Testing [ 10002 ] | Post-merge Testing In Progress [ 10003 ] |
| Resolution | Done [ 10000 ] | |
| Status | Post-merge Testing In Progress [ 10003 ] | Closed [ 6 ] |
| Story Points | 1 | 3 |
| Assignee | Paige Kulzer [ pkulzer ] | Ann Loraine [ aloraine ] |
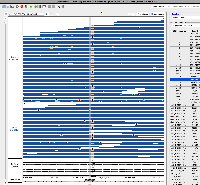
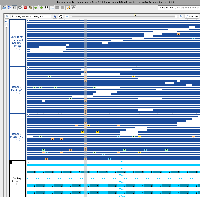
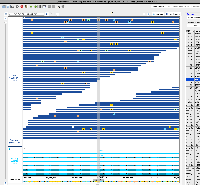
I've confirmed that I can visualize this CRAM data in IGB 10.1.0 (although the VCF file doesn't seem to be loading). Although it's unclear if Nebula Genomics was the genome re-sequencing company that produced this data, I think we should move forward with deploying it and making it available as an IGB Quickload ahead of the ASHG conference this Fall.
Passing this ticket to Nowlan Freese for next steps.