From: Mason Meyer <mmeyer20@uncc.edu>
To: Rajesh Biswas <biswasr0@gmail.com>
CC: "Loraine, Ann" <Ann.Loraine@uncc.edu>
Date: Wed, Sep 20, 2017 at 10:26 AM
Subject: Re: view read sequence
Hello Rajesh,
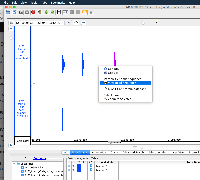
My name is Mason Meyer, IGB Support Specialist. Thank you for contacting us with your question. After viewing the screenshots you sent, it appears to me that your files are BED files and the "View Read Sequence" feature will only work for BAM files containing read alignments.
For your case, the "View Genomic Sequence" option should allow you to view the selected peak's sequence. If that allows you to accomplish your goal, please let me know and I will mark your issue as resolved, but if not, please don't hesitate to contact me and I will continue to look into the issue. Also, if you have any additional feedback for our team, we would love to hear it!
Thanks again,
Mason Meyer
IGB Support Specialist
Bug
![]()
From: Mason Meyer <mmeyer20@uncc.edu>
To: Rajesh Biswas <biswasr0@gmail.com>
CC: "Loraine, Ann" <Ann.Loraine@uncc.edu>
Date: Wed, Sep 20, 2017 at 10:26 AM
Subject: Re: view read sequence
Hello Rajesh,
My name is Mason Meyer, IGB Support Specialist. Thank you for contacting us with your question. After viewing the screenshots you sent, it appears to me that your files are BED files and the "View Read Sequence" feature will only work for BAM files containing read alignments.
For your case, the "View Genomic Sequence" option should allow you to view the selected peak's sequence. If that allows you to accomplish your goal, please let me know and I will mark your issue as resolved, but if not, please don't hesitate to contact me and I will continue to look into the issue. Also, if you have any additional feedback for our team, we would love to hear it!
Thanks again,
Mason Meyer
IGB Support Specialist