From: Mason Meyer <mmeyer20@uncc.edu>
To: Colleen Bianco <cbianco2@illinois.edu>
Date: Thu, Nov 30, 2017 at 3:30 PM
Subject: Re: Trouble loading custom reference genome
Hello Colleen,
My name is Mason Meyer, IGB Support Specialist. Thank you for contacting us for assistance!
I apologize for the delay in my response. I actually thought I had e-mailed you back several days ago, but I just realized there was no recipient set in the e-mail! Only my teammates were CC'd on the e-mail. I apologize for this mistake!
I looked into your issue and tried to identify the Genbank number for the E. coli reference genome in IGB, but I was not able to find it. However, I was able to find a Genbank and FASTA file for the Escherichia coli str. K-12 substr. MG1655, complete genome, GenBank: U00096.3. I was able to download them here:
https://www.ncbi.nlm.nih.gov/nuccore/U00096.3
You may have already downloaded those files, but for your convenience, I am attaching them to this e-mail, along with a chromosome.txt file which is needed for IGB to recognize that the files are using the same chromosomes.
To load the chromosome.txt file, please follow the steps below:
Inline image 1
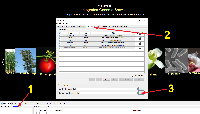
1) Start IGB and click the Configure text in the lower-left corner of IGB to open the IGB Preferences Panel (see screenshot).
2) Select the Data Sources tab in the IGB Preferences Panel.
3) Load the chromosome.txt file by clicking on the "..." button for the Chromosome Synonyms File and selecting your file using the file browser. Once, you have selected the chromosome.txt file, close the IGB Preferences Panel.
After that, you can load the .gb and .fasta files into IGB. The quickest and easiest way to open these files into IGB is to drag-and drop the .gb and .fasta files right onto the IGB window before selecting a genome. Once IGB opens the files as new tracks, you can click the Load Data button to load the data in view.
With these files and instructions, you should be able to visualize your desired genome, as expected. If you run into any issues or have any questions, please let me know. I would also love to know if my assistance was able to help you resolve the issue. I look forward to hearing back from you!
Thanks again,
Mason Meyer
IGB Support Specialist
Bug
Blocker
![]()
From: Mason Meyer <mmeyer20@uncc.edu>
To: Colleen Bianco <cbianco2@illinois.edu>
Date: Thu, Nov 30, 2017 at 3:30 PM
Subject: Re: Trouble loading custom reference genome
Hello Colleen,
My name is Mason Meyer, IGB Support Specialist. Thank you for contacting us for assistance!
I apologize for the delay in my response. I actually thought I had e-mailed you back several days ago, but I just realized there was no recipient set in the e-mail! Only my teammates were CC'd on the e-mail. I apologize for this mistake!
I looked into your issue and tried to identify the Genbank number for the E. coli reference genome in IGB, but I was not able to find it. However, I was able to find a Genbank and FASTA file for the Escherichia coli str. K-12 substr. MG1655, complete genome, GenBank: U00096.3. I was able to download them here:
https://www.ncbi.nlm.nih.gov/nuccore/U00096.3
You may have already downloaded those files, but for your convenience, I am attaching them to this e-mail, along with a chromosome.txt file which is needed for IGB to recognize that the files are using the same chromosomes.
To load the chromosome.txt file, please follow the steps below:
Inline image 1
1) Start IGB and click the Configure text in the lower-left corner of IGB to open the IGB Preferences Panel (see screenshot).
2) Select the Data Sources tab in the IGB Preferences Panel.
3) Load the chromosome.txt file by clicking on the "..." button for the Chromosome Synonyms File and selecting your file using the file browser. Once, you have selected the chromosome.txt file, close the IGB Preferences Panel.
After that, you can load the .gb and .fasta files into IGB. The quickest and easiest way to open these files into IGB is to drag-and drop the .gb and .fasta files right onto the IGB window before selecting a genome. Once IGB opens the files as new tracks, you can click the Load Data button to load the data in view.
With these files and instructions, you should be able to visualize your desired genome, as expected. If you run into any issues or have any questions, please let me know. I would also love to know if my assistance was able to help you resolve the issue. I look forward to hearing back from you!
Thanks again,
Mason Meyer
IGB Support Specialist