Details
-
Type:
Bug
-
Status: Closed (View Workflow)
-
Priority:
Blocker
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: 9.1.0 Major Release
-
Labels:
-
Story Points:2
-
Epic Link:
-
Sprint:Winter 2018 Sprint 3, Fall 2019 Sprint 1, Fall 2019 Sprint 2, Fall 4 : 30 Sep to 11 Oct, Fall 5 : 14 Oct to 25 Oct, Fall 6 : 28 Oct to 8 Nov
Description
Working with PacBio long reads from a URL, hit a SocketTimeoutException. The reads do not display correctly - appearing as unfilled squares. The read scores also seem to be incorrect, though that may be a separate issue.
Example bam can be found here
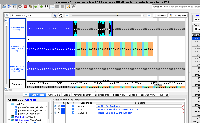
This issue may not have anything to do with the reads themselves, and may be an issue with the data being pulled from an ftp site. However, when I pull Illumina short reads from the same site, they load in IGB normally. We haven't tested long reads very much in IGB (PacBio are around 10,000 bp long, versus Illumina 150 bp long), and I'm wondering if the long reads are behaving correctly.