Details
-
Type:
Improvement
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: 9.1.10 Major Release
-
Labels:None
-
Story Points:4
-
Epic Link:
-
Sprint:Fall 6 2021 Oct 25 - Nov 5, Fall 7 2021 Nov 8 - Nov 24, Fall 8 2021 Nov 29 - Dec 10, Fall 9 2021 Dec 13 - Dec 24, Spring 1 2022 Jan 3 - Jan 14
Description
Situation: bigBed is an indexed binary format of bed files. However, in IGB, bigBed files are not appearing correctly. The bigBed gene annotations (see attached file) are not showing the introns/exons, but instead are drawing a single glyph for the entire gene.
Task: Fix IGB so that the bigBed files appear correctly.
See the UCSC guide on bigBed for more information on the bigBed file format.
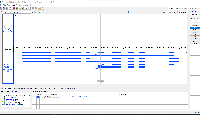

To reproduce the issue:
Open IGB
Open the Arabidopsis thaliana A_thaliana_Jun_2009 genome
Navigate to Chr1:2,257,253-2,260,326
Load the attached Araport11.bb file
Click Load Data and compare the Araport11.bb file and the Araport11 bed file found in IGB Quickload (should load by default)