Details
-
Type:
New Feature
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: None
-
Labels:None
-
Story Points:1
-
Epic Link:
-
Sprint:Fall 7 2021 Nov 8 - Nov 24, Fall 8 2021 Nov 29 - Dec 10, Fall 9 2021 Dec 13 - Dec 24
Description
A method for determining whether or not a UCSC genome name can be mapped to an IGB genome version is needed in order to decide which genome versions in the trackhub table UI can be made to have the option of utilizing IGB's 'open genome' endpoint located at http://localhost:7085/IGBControl?version=GENOME. Since this functionality may be needed in future services, an endpoint can be added to the trackhub facade API that returns the primary IGB genome version name corresponding to the provided UCSC genome version. This can be accomplished by searching the master synonyms.txt in an attempt to find an entry that matches the UCSC genome name. The first entry of the corresponding row would then be returned as the primary IGB genome version name.
It may be useful to extend IGB's endpoint to also include an 'species' query parameter, which could allow users to open converted trackhubs whose genome versions do not map to those associated with an IGB-supported organism.
Attachments
Issue Links
Activity
| Field | Original Value | New Value |
|---|---|---|
| Epic Link | IGBF-2831 [ 19524 ] |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Description | A method for determining whether or not a UCSC genome name can be mapped to an IGB genome version is needed in order to decide which genome versions in the trackhub table UI can be made to have the option of utilizing IGB's 'open genome' endpoint located at http://localhost:7085/IGBControl?version=${genome}. Since this functionality may be needed in future services, an endpoint can be added to the trackhub facade API that returns the primary IGB genome version name corresponding to the provided UCSC genome version. This can be accomplished by searching the master [synonyms.txt|https://bitbucket.org/lorainelab/integrated-genome-browser/raw/3ea35fe2b8fd3c9dc59c1f3390744b41461f90ae/core/synonym-lookup/src/main/resources/synonyms.txt] in an attempt to find an entry that matches the UCSC genome name. The first entry of the corresponding row would then be returned as the primary IGB genome version name. |
| Description | A method for determining whether or not a UCSC genome name can be mapped to an IGB genome version is needed in order to decide which genome versions in the trackhub table UI can be made to have the option of utilizing IGB's 'open genome' endpoint located at http://localhost:7085/IGBControl?version=${genome}. Since this functionality may be needed in future services, an endpoint can be added to the trackhub facade API that returns the primary IGB genome version name corresponding to the provided UCSC genome version. This can be accomplished by searching the master [synonyms.txt|https://bitbucket.org/lorainelab/integrated-genome-browser/raw/3ea35fe2b8fd3c9dc59c1f3390744b41461f90ae/core/synonym-lookup/src/main/resources/synonyms.txt] in an attempt to find an entry that matches the UCSC genome name. The first entry of the corresponding row would then be returned as the primary IGB genome version name. | A method for determining whether or not a UCSC genome name can be mapped to an IGB genome version is needed in order to decide which genome versions in the trackhub table UI can be made to have the option of utilizing IGB's 'open genome' endpoint located at http://localhost:7085/IGBControl?version=GENOME. Since this functionality may be needed in future services, an endpoint can be added to the trackhub facade API that returns the primary IGB genome version name corresponding to the provided UCSC genome version. This can be accomplished by searching the master [synonyms.txt|https://bitbucket.org/lorainelab/integrated-genome-browser/raw/3ea35fe2b8fd3c9dc59c1f3390744b41461f90ae/core/synonym-lookup/src/main/resources/synonyms.txt] in an attempt to find an entry that matches the UCSC genome name. The first entry of the corresponding row would then be returned as the primary IGB genome version name. |
| Description | A method for determining whether or not a UCSC genome name can be mapped to an IGB genome version is needed in order to decide which genome versions in the trackhub table UI can be made to have the option of utilizing IGB's 'open genome' endpoint located at http://localhost:7085/IGBControl?version=GENOME. Since this functionality may be needed in future services, an endpoint can be added to the trackhub facade API that returns the primary IGB genome version name corresponding to the provided UCSC genome version. This can be accomplished by searching the master [synonyms.txt|https://bitbucket.org/lorainelab/integrated-genome-browser/raw/3ea35fe2b8fd3c9dc59c1f3390744b41461f90ae/core/synonym-lookup/src/main/resources/synonyms.txt] in an attempt to find an entry that matches the UCSC genome name. The first entry of the corresponding row would then be returned as the primary IGB genome version name. |
A method for determining whether or not a UCSC genome name can be mapped to an IGB genome version is needed in order to decide which genome versions in the trackhub table UI can be made to have the option of utilizing IGB's 'open genome' endpoint located at http://localhost:7085/IGBControl?version=GENOME. Since this functionality may be needed in future services, an endpoint can be added to the trackhub facade API that returns the primary IGB genome version name corresponding to the provided UCSC genome version. This can be accomplished by searching the master [synonyms.txt|https://bitbucket.org/lorainelab/integrated-genome-browser/raw/3ea35fe2b8fd3c9dc59c1f3390744b41461f90ae/core/synonym-lookup/src/main/resources/synonyms.txt] in an attempt to find an entry that matches the UCSC genome name. The first entry of the corresponding row would then be returned as the primary IGB genome version name.
It may be useful to extend IGB's endpoint to also include an 'species' query parameter, which could allow users to open converted trackhubs whose genome versions do not map to those associated with an IGB-supported organism. |
| Sprint | Fall 7 2021 Nov 8 - Nov 24 [ 133 ] | Fall 7 2021 Nov 8 - Nov 24, Fall 8 2021 Nov 29 - Dec 10 [ 133, 134 ] |
| Rank | Ranked higher |
| Status | In Progress [ 3 ] | To-Do [ 10305 ] |
| Summary | Add trackhub facade API endpoint to check for UCSC-IGB genome version match | Add trackhub facade API endpoint to translate UCSC genome names to IGB synonym equivalents |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Status | In Progress [ 3 ] | Needs 1st Level Review [ 10005 ] |
| Assignee | Philip Badzuh [ pbadzuh ] |
| Assignee | Ann Loraine [ aloraine ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Status | First Level Review in Progress [ 10301 ] | Ready for Pull Request [ 10304 ] |
| Assignee | Ann Loraine [ aloraine ] | Philip Badzuh [ pbadzuh ] |
| Status | Ready for Pull Request [ 10304 ] | Pull Request Submitted [ 10101 ] |
| Assignee | Philip Badzuh [ pbadzuh ] |
| Assignee | Philip Badzuh [ pbadzuh ] |
| Status | Pull Request Submitted [ 10101 ] | Reviewing Pull Request [ 10303 ] |
| Status | Reviewing Pull Request [ 10303 ] | To-Do [ 10305 ] |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Status | In Progress [ 3 ] | Needs 1st Level Review [ 10005 ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Status | First Level Review in Progress [ 10301 ] | Ready for Pull Request [ 10304 ] |
| Status | Ready for Pull Request [ 10304 ] | Pull Request Submitted [ 10101 ] |
| Assignee | Philip Badzuh [ pbadzuh ] |
| Status | Pull Request Submitted [ 10101 ] | Reviewing Pull Request [ 10303 ] |
| Status | Reviewing Pull Request [ 10303 ] | Merged Needs Testing [ 10002 ] |
| Assignee | Omkar Marne [ omarne ] |
| Sprint | Fall 7 2021 Nov 8 - Nov 24, Fall 8 2021 Nov 29 - Dec 10 [ 133, 134 ] | Fall 7 2021 Nov 8 - Nov 24, Fall 8 2021 Nov 29 - Dec 10, Fall 9 2021 Dec 13 - Dec 24 [ 133, 134, 135 ] |
| Rank | Ranked higher |
| Attachment | screenshot-1.png [ 17020 ] |
| Status | Merged Needs Testing [ 10002 ] | Post-merge Testing In Progress [ 10003 ] |
| Status | Post-merge Testing In Progress [ 10003 ] | To-Do [ 10305 ] |
| Assignee | Omkar Marne [ omarne ] | Philip Badzuh [ pbadzuh ] |
| Assignee | Philip Badzuh [ pbadzuh ] | Omkar Marne [ omarne ] |
| Attachment | Postman.png [ 17021 ] |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Status | In Progress [ 3 ] | Needs 1st Level Review [ 10005 ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Status | First Level Review in Progress [ 10301 ] | Ready for Pull Request [ 10304 ] |
| Status | Ready for Pull Request [ 10304 ] | Pull Request Submitted [ 10101 ] |
| Status | Pull Request Submitted [ 10101 ] | Reviewing Pull Request [ 10303 ] |
| Status | Reviewing Pull Request [ 10303 ] | Merged Needs Testing [ 10002 ] |
| Status | Merged Needs Testing [ 10002 ] | Post-merge Testing In Progress [ 10003 ] |
| Assignee | Omkar Marne [ omarne ] | Philip Badzuh [ pbadzuh ] |
| Status | Post-merge Testing In Progress [ 10003 ] | To-Do [ 10305 ] |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Status | In Progress [ 3 ] | Needs 1st Level Review [ 10005 ] |
| Assignee | Philip Badzuh [ pbadzuh ] |
| Assignee | Omkar Marne [ omarne ] |
| Assignee | Omkar Marne [ omarne ] | Ann Loraine [ aloraine ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Status | First Level Review in Progress [ 10301 ] | Ready for Pull Request [ 10304 ] |
| Assignee | Ann Loraine [ aloraine ] | Philip Badzuh [ pbadzuh ] |
| Status | Ready for Pull Request [ 10304 ] | Pull Request Submitted [ 10101 ] |
| Assignee | Philip Badzuh [ pbadzuh ] |
| Status | Pull Request Submitted [ 10101 ] | Reviewing Pull Request [ 10303 ] |
| Status | Reviewing Pull Request [ 10303 ] | Merged Needs Testing [ 10002 ] |
| Assignee | Omkar Marne [ omarne ] |
| Attachment | screenshot-2.png [ 17023 ] |
| Attachment | screenshot-1.png [ 17020 ] |
| Status | Merged Needs Testing [ 10002 ] | Post-merge Testing In Progress [ 10003 ] |
| Resolution | Done [ 10000 ] | |
| Status | Post-merge Testing In Progress [ 10003 ] | Closed [ 6 ] |
| Assignee | Omkar Marne [ omarne ] | Philip Badzuh [ pbadzuh ] |
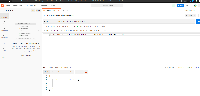
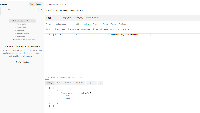
I started by implementing the endpoint to take a single UCSC genome and return the IGB equivalent, however, this would require many requests that would increase latency for the front-end.
As a second version, I updated the endpoint to take multiple UCSC genome versions for conversion.
In testing, there was a CORS issue, so I configured our django server to allow requests from all origins in the browser. We can update this to limit it to bioviz.org, if you see this as an issue, [~aloraine].