Details
-
Type:
Task
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: None
-
Labels:None
-
Story Points:0.5
-
Epic Link:
-
Sprint:Spring 2 2022 Jan 18 - Jan 28
Description
University of Arizona researcher Dr. Cedar Warman is comparing data collected from two different library synthesis processes in which samples prepared by the U. of Az lab were compared to libraries made by a company - GeneWiz, combined, and then sequenced in a single lane.
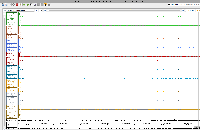
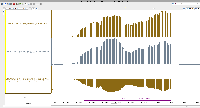
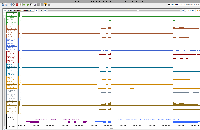
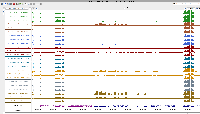
To analyze results, he has created genome coverage graphs using bamCoverage (deepTools).
For this task, we will set up an on-line call to show relevant features in IGB, mainly graph manipulation methods, that could help him analyze his data. During the zoom call, we will try to highlight ways that visualization can expose interesting features in data and even suggest better ways to analyze the data.
Note that there are a million and one ways to perform statistical analysis of data, i.e., there are many models that can be used. Selecting the right model for one's own data requires data exploration, to gain understanding of one's data and to notice salient properties. For genomic data, one of the most effective strategies for noticing salient features is to interactively view and explore the data in a genome browser such as IGB.
Attachments
Issue Links
- relates to
-
IGBF-3070 Replicate graph addition problem
-
- Closed
-