Details
-
Type:
Bug
-
Status: To-Do (View Workflow)
-
Priority:
Blocker
-
Resolution: Unresolved
-
Labels:
Description
When I load an annotation file (gff3) using the Windows version of IGB, it does not show the option to show the gene name (gene=) from the gff3 file (as seen in the drop down menu). If I use this same file in the Linux version, it gives me the right option and a few more.
I would like to be able to label by gene name eg. "lysT" instead of this UNKNOWN_SYM65 id.
GFF file in this example region:
AL123456 EMBL gene 923803 923875 . - . ID=gene870;Name=lysT;gbkey=Gene;gene=lysT;gene_biotype=tRNA
AL123456 EMBL tRNA 923803 923875 . - . ID=rna13;Parent=gene870;Note=codon recognized: AAA - lysT - tRNA-Lys - anticodon ttt - length %3D 73;anticodon=(pos:complement(923840..923842));gbkey=tRNA;gene=lysT;product=tRNA-Lys
AL123456 EMBL exon 923803 923875 . - . ID=id50;Parent=rna13;Note=codon recognized: AAA - lysT - tRNA-Lys - anticodon ttt - length %3D 73;anticodon=(pos:complement(923840..923842));gbkey=tRNA;gene=lysT;product=tRNA-Lys
Reporter: Valdir Barth
E-mail: valdirbarth@gmail.com![]()
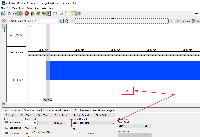
From: Mason Meyer <mmeyer20@uncc.edu>
To: Valdir Barth <valdirbarth@gmail.com>
CC: Ann Loraine <Ann.Loraine@uncc.edu>
Date: Mon, Oct 30, 2017 at 6:58 PM
Subject: Re: Windows version does not read gff file properly
Hello Valdir,
My name is Mason Meyer, IGB Support Specialist. Thanks for contacting our team for assistance with your issue.
I spent some time trying to reproduce the issue of the gene label not displaying on Windows, but I was not yet able to. When trying to reproduce the issue on my Windows machine, I was able to label by the gene field so that the text "lysT" was displayed (see screenshot). When I copied the data from your e-mail, I did have to convert the spaces into tabs so IGB would open the file, but I imagine you didn't experience this problem since you have the actual file.
I have been trying to figure out what might be causing this label option issue for you and not me. Is your file labeled with the extension ".gff3"?
I feel confident that we can resolve the issue quickly if we figure out why it is happening. Are you able to send me the GFF3 file to use for testing? If not, please let me know and we can look into other ways of resolving the issue.
Thanks again,
Mason Meyer
IGB Support Specialist