Details
-
Type:
Task
-
Status: Closed (View Workflow)
-
Priority:
Major
-
Resolution: Done
-
Affects Version/s: None
-
Fix Version/s: None
-
Labels:None
-
Story Points:0.5
-
Epic Link:
-
Sprint:Spring 2 2022 Jan 18 - Jan 28, Spring 3 2022 Jan 31 - Feb 11, Spring 4 2022 Feb 14 - Feb 25
Description
During a live demo, one of the attendees noticed a problem with a graph operation.
The sum of two graphs did not appear to be correct.
To repeat / investigate:
- Add data source from
IGBF-3064 - Load six scaled coverage graphs from the graphs folder
- Select 3 of the graphs by shift-clicking the left side graph track labels (red outline appears around the label when the graph track is selected)
- Select use the Graph Operations multi-graph menu "Mean" to create a new graph, which is supposed to be the average of the selected graphs
- Repeat the above with another 3 graphs
- Now, subtract the mean graphs from each other
- Check the y-axis values carefully to ensure that the subtraction was correctly done.
Report what you see, along with images showing incorrect or correct behavior.
Attachments
Issue Links
Activity
| Field | Original Value | New Value |
|---|---|---|
| Epic Link | IGBF-1765 [ 17855 ] |
| Description |
During a live demo, one of the attendees noticed a problem with a graph operation.
The sum of two graphs did not appear to be correct. |
During a live demo, one of the attendees noticed a problem with a graph operation.
The sum of two graphs did not appear to be correct. To repeat / investigate: * Add data source from * Load six scaled coverage graphs from the graphs folder * Select 3 of the graphs by shift-clicking the left side graph track labels (red outline appears around the label when the graph track is selected) * Select use the Graph Operations multi-graph menu "Mean" to create a new graph, which is supposed to be the average of the selected graphs * Repeat the above with another 3 graphs * Now, subtract the mean graphs from each other * Check the y-axis values carefully to ensure that the subtraction was correctly done. Report what you see, along with images showing incorrect or correct behavior. |
| Assignee | Omkar Marne [ omarne ] |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Attachment | Mean.png [ 17086 ] |
| Attachment | Subtract.png [ 17087 ] |
| Attachment | Subtract.png [ 17088 ] |
| Status | In Progress [ 3 ] | Needs 1st Level Review [ 10005 ] |
| Assignee | Omkar Marne [ omarne ] | Ann Loraine [ aloraine ] |
| Attachment | Addition.png [ 17089 ] |
| Sprint | Spring 2 2022 Jan 18 - Jan 28 [ 137 ] | Spring 2 2022 Jan 18 - Jan 28, Spring 3 2022 Jan 31 - Feb 11 [ 137, 138 ] |
| Rank | Ranked higher |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Status | First Level Review in Progress [ 10301 ] | To-Do [ 10305 ] |
| Assignee | Ann Loraine [ aloraine ] |
| Assignee | Omkar Marne [ omarne ] |
| Assignee | Omkar Marne [ omarne ] |
| Assignee | Omkar Marne [ omarne ] |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Sprint | Spring 2 2022 Jan 18 - Jan 28, Spring 3 2022 Jan 31 - Feb 11 [ 137, 138 ] | Spring 2 2022 Jan 18 - Jan 28, Spring 3 2022 Jan 31 - Feb 11, Spring 4 2022 Feb 14 - Feb 25 [ 137, 138, 139 ] |
| Rank | Ranked higher |
| Attachment | Addition.png [ 17089 ] |
| Attachment | Mean.png [ 17086 ] |
| Attachment | Subtract.png [ 17088 ] |
| Attachment | Subtract.png [ 17087 ] |
| Attachment | y-axis - 1st graph 4085.7.png [ 17176 ] |
| Attachment | y-axis 2nd graph 3389.1.png [ 17177 ] |
| Attachment | y-axis 3rd graph 3707.9.png [ 17178 ] |
| Attachment | Mean 1st,2nd,3rd 3726.7.png [ 17179 ] |
| Attachment | 4th graph 3499.4.png [ 17180 ] |
| Attachment | 5th graph 4167.3.png [ 17181 ] |
| Attachment | 6th graph 4369.3.png [ 17182 ] |
| Attachment | Mean 4th,5th,6th - 4012.png [ 17183 ] |
| Attachment | Mean difference.png [ 17184 ] |
| Comment | [ Mean result, addition result and subtraction result is correct for the selected graphs. Please see attached images. ] |
| Attachment | Subtraction.png [ 17185 ] |
| Attachment | 4th graph 3499.4.png [ 17180 ] |
| Attachment | 5th graph 4167.3.png [ 17181 ] |
| Attachment | 6th graph 4369.3.png [ 17182 ] |
| Attachment | Mean 1st,2nd,3rd 3726.7.png [ 17179 ] |
| Attachment | Mean 4th,5th,6th - 4012.png [ 17183 ] |
| Attachment | Mean difference.png [ 17184 ] |
| Attachment | Subtraction.png [ 17185 ] |
| Attachment | y-axis - 1st graph 4085.7.png [ 17176 ] |
| Attachment | y-axis 2nd graph 3389.1.png [ 17177 ] |
| Attachment | y-axis 3rd graph 3707.9.png [ 17178 ] |
| Attachment | Mean 1.png [ 17186 ] |
| Attachment | Mean 1.png [ 17186 ] |
| Attachment | Mean 1.png [ 17187 ] |
| Attachment | Mean 2.png [ 17188 ] |
| Attachment | Difference.png [ 17189 ] |
| Status | In Progress [ 3 ] | Needs 1st Level Review [ 10005 ] |
| Assignee | Omkar Marne [ omarne ] | Ann Loraine [ aloraine ] |
| Assignee | Ann Loraine [ aloraine ] |
| Assignee | Nowlan Freese [ nfreese ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Attachment | 48738118.png [ 17190 ] |
| Assignee | Nowlan Freese [ nfreese ] |
| Status | First Level Review in Progress [ 10301 ] | Needs 1st Level Review [ 10005 ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Status | First Level Review in Progress [ 10301 ] | Needs 1st Level Review [ 10005 ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Assignee | Nowlan Freese [ nfreese ] |
| Status | First Level Review in Progress [ 10301 ] | Needs 1st Level Review [ 10005 ] |
| Assignee | Nowlan Freese [ nfreese ] | Ann Loraine [ aloraine ] |
| Assignee | Ann Loraine [ aloraine ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Status | First Level Review in Progress [ 10301 ] | To-Do [ 10305 ] |
| Status | To-Do [ 10305 ] | In Progress [ 3 ] |
| Status | In Progress [ 3 ] | Needs 1st Level Review [ 10005 ] |
| Status | Needs 1st Level Review [ 10005 ] | First Level Review in Progress [ 10301 ] |
| Status | First Level Review in Progress [ 10301 ] | Ready for Pull Request [ 10304 ] |
| Status | Ready for Pull Request [ 10304 ] | Pull Request Submitted [ 10101 ] |
| Status | Pull Request Submitted [ 10101 ] | Reviewing Pull Request [ 10303 ] |
| Status | Reviewing Pull Request [ 10303 ] | Merged Needs Testing [ 10002 ] |
| Status | Merged Needs Testing [ 10002 ] | Post-merge Testing In Progress [ 10003 ] |
| Resolution | Done [ 10000 ] | |
| Status | Post-merge Testing In Progress [ 10003 ] | Closed [ 6 ] |
| Assignee | Omkar Marne [ omarne ] |
| Summary | Investigate graph addition problem | Replicate graph addition problem |

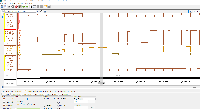
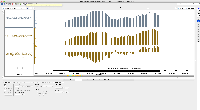
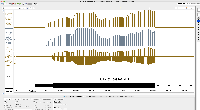
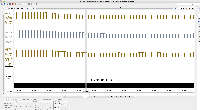
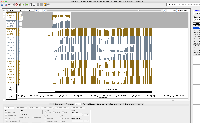
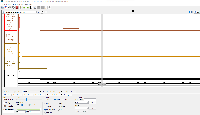
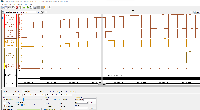
I am not seeing evidence in the images of correct values. The view is too zoomed-out to investigate individual values at the individual base pair level.
For next step, we need to look more closely at image file scaledcoveragegraphs-ARE-chr1-diff-problem.png from the linked ticket
IGBF-3064.Please load the same scene and investigate. Do the y-axis values make sense? Check the "diff" graph track, which is supposed to be the subtraction of one computed mean track minus the other.